Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis
- PMID: 18323469
- PMCID: PMC2409274
- DOI: 10.1210/me.2007-0565
Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis
Abstract
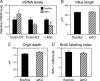
Liver receptor homolog 1 (LRH-1), an orphan nuclear receptor, is highly expressed in liver and intestine, where it is implicated in the regulation of cholesterol, bile acid, and steroid hormone homeostasis. Among the proposed LRH-1 target genes in liver are those encoding cholesterol 7alpha-hydroxylase (CYP7A1) and sterol 12alpha-hydroxylase (CYP8B1), which catalyze key steps in bile acid synthesis. In vitro studies suggest that LRH-1 may be involved both in stimulating basal CYP7A1 and CYP8B1 transcription and in repressing their expression as part of the nuclear bile acid receptor [farnesoid X receptor (FXR)]-small heterodimer partner signaling cascade, which culminates in small heterodimer partner binding to LRH-1 to repress gene transcription. However, in vivo analysis of LRH-1 actions has been hampered by the embryonic lethality of Lrh-1 knockout mice. To overcome this obstacle, mice were generated in which Lrh-1 was selectively disrupted in either hepatocytes or intestinal epithelium. LRH-1 deficiency in either tissue changed mRNA levels of genes involved in cholesterol and bile acid homeostasis. Surprisingly, LRH-1 deficiency in hepatocytes had no significant effect on basal Cyp7a1 expression or its repression by FXR. Whereas Cyp8b1 repression by FXR was also intact in mice deficient for LRH-1 in hepatocytes, basal CYP8B1 mRNA levels were significantly decreased, and there were corresponding changes in the composition of the bile acid pool. Taken together, these data reveal a broad role for LRH-1 in regulating bile acid homeostasis but demonstrate that LRH-1 is either not involved in the feedback regulation of bile acid synthesis or is compensated for by other factors.
Figures



References
-
- Fayard E, Auwerx J, Schoonjans K 2004 LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol 14:250–260 - PubMed
-
- Lee YK, Choi YH, Chua S, Park YJ, Moore DD 2006 Phosphorylation of the hinge domain of the nuclear hormone receptor LRH-1 stimulates transactivation. J Biol Chem 281:7850–7855 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

