Spatial separation of Golgi and ER during mitosis protects SREBP from unregulated activation
- PMID: 18323777
- PMCID: PMC2323267
- DOI: 10.1038/emboj.2008.36
Spatial separation of Golgi and ER during mitosis protects SREBP from unregulated activation
Abstract
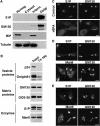
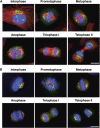
Sterol regulatory element-binding proteins (SREBPs) are membrane-bound transcription factors that reside as inactive precursors in the endoplasmic reticulum (ER) membrane. After sterol depletion, the proteins are transported to the Golgi apparatus, where they are cleaved by site-1 protease (S1P). Cleavage releases the active transcription factors, which then enter the nucleus to induce genes that regulate cellular levels of cholesterol and phospholipids. This regulation depends on the spatial separation of the Golgi and the ER, as mixing of the compartments induces unregulated activation of SREBPs. Here, we show that S1P is localized to the Golgi, but cycles continuously through the ER and becomes trapped when ER exit is inhibited. During mitosis, S1P is associated with mitotic Golgi clusters, which remain distinct from the ER. In mitotic cells, S1P is active, but SREBP is not cleaved as S1P and SREBP reside in different compartments. Together, these results indicate that the spatial separation of the Golgi and the ER is maintained during mitosis, which is essential to protect the S1P substrate SREBP from unregulated activation during mitosis.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

