Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter
- PMID: 18326675
- PMCID: PMC2394895
- DOI: 10.1128/AEM.02311-07
Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter
Abstract
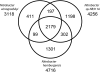
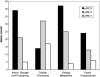
The alphaproteobacterium Nitrobacter hamburgensis X14 is a gram-negative facultative chemolithoautotroph that conserves energy from the oxidation of nitrite to nitrate. Sequencing and analysis of the Nitrobacter hamburgensis X14 genome revealed four replicons comprised of one chromosome (4.4 Mbp) and three plasmids (294, 188, and 121 kbp). Over 20% of the genome is composed of pseudogenes and paralogs. Whole-genome comparisons were conducted between N. hamburgensis and the finished and draft genome sequences of Nitrobacter winogradskyi and Nitrobacter sp. strain Nb-311A, respectively. Most of the plasmid-borne genes were unique to N. hamburgensis and encode a variety of functions (central metabolism, energy conservation, conjugation, and heavy metal resistance), yet approximately 21 kb of a approximately 28-kb "autotrophic" island on the largest plasmid was conserved in the chromosomes of Nitrobacter winogradskyi Nb-255 and Nitrobacter sp. strain Nb-311A. The N. hamburgensis chromosome also harbors many unique genes, including those for heme-copper oxidases, cytochrome b(561), and putative pathways for the catabolism of aromatic, organic, and one-carbon compounds, which help verify and extend its mixotrophic potential. A Nitrobacter "subcore" genome was also constructed by removing homologs found in strains of the closest evolutionary relatives, Bradyrhizobium japonicum and Rhodopseudomonas palustris. Among the Nitrobacter subcore inventory (116 genes), copies of genes or gene clusters for nitrite oxidoreductase (NXR), cytochromes associated with a dissimilatory nitrite reductase (NirK), PII-like regulators, and polysaccharide formation were identified. Many of the subcore genes have diverged significantly from, or have origins outside, the alphaproteobacterial lineage and may indicate some of the unique genetic requirements for nitrite oxidation in Nitrobacter.
Figures

References
-
- Ahlers, B., W. Konig, and E. Bock. 1990. Nitrite reductase activity in Nitrobacter vulgaris. FEMS Microbiol. Lett. 67:121-126.
-
- Arkin, I. T., H. Xu, M. O. Jensen, E. Arbely, E. R. Bennett, K. J. Bowers, E. Chow, R. O. Dror, M. P. Eastwood, R. Flitman-Tene, B. A. Gregersen, J. L. Klepeis, I. Kolossvary, Y. Shan, and D. E. Shaw. 2007. Mechanism of Na+/H+ antiporting. Science 317:799-803. - PubMed
-
- Bock, E. 1976. Growth of Nitrobacter in the presence of organic matter. II. Chemoorganotrophic growth of Nitrobacter agilis. Arch. Microbiol. 108:305-312. - PubMed
-
- Bock, E., H. P. Koops, H. Harms, and B. Ahlers. 1991. The biochemistry of nitrifying organisms, p. 171-200. In J. M. Shively and L. L. Barton (ed.), Variations in autotrophic life. Academic Press, San Diego, CA.
-
- Bock, E., H.-P. Koops, U. C. Möller, and M. Rudert. 1990. A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch. Microbiol. 153:105-110.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

