Mechanism for oxidation of high-molecular-weight substrates by a fungal versatile peroxidase, MnP2
- PMID: 18326680
- PMCID: PMC2394877
- DOI: 10.1128/AEM.02080-07
Mechanism for oxidation of high-molecular-weight substrates by a fungal versatile peroxidase, MnP2
Abstract
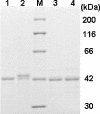
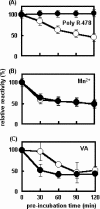
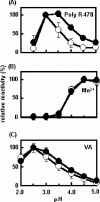
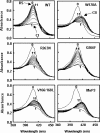
Unlike general peroxidases, Pleurotus ostreatus MnP2 was reported to have a unique property of direct oxidization of high-molecular-weight compounds, such as Poly R-478 and RNase A. To elucidate the mechanism for oxidation of polymeric substrates by MnP2, a series of mutant enzymes were produced by using a homologous gene expression system, and their reactivities were characterized. A mutant enzyme with an Ala substituting for an exposing Trp (W170A) drastically lost oxidation activity for veratryl alcohol (VA), Poly R-478, and RNase A, whereas the kinetic properties for Mn(2+) and H(2)O(2) were substantially unchanged. These results demonstrated that, in addition to VA, the high-molecular-weight substrates are directly oxidized by MnP2 at W170. Moreover, in the mutants Q266F and V166/168L, amino acid substitution(s) around W170 resulted in a decreased activity only for the high-molecular-weight substrates. These results, along with the three-dimensional modeling of the mutants, suggested that the mutations caused a steric hindrance to access of the polymeric substrates to W170. Another mutant, R263N, contained a newly generated N glycosylation site and showed a higher molecular mass in sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. Interestingly, the R263N mutant exhibited an increased reactivity with VA and high-molecular-weight substrates. The existence of an additional carbohydrate modification and the catalytic properties in this mutant are discussed. This is the first study of a direct mechanism for oxidation of high-molecular-weight substrates by a fungal peroxidase using a homologous gene expression system.
Figures


References
-
- Amado, R., R. Aeschbach, and H. Neukom. 1984. Dityrosine: in vitro production and characterization. Methods Enzymol. 107:377-388. - PubMed
-
- Asada, Y., A. Watanabe, T. Irie, T. Nakayama, and M. Kuwahara. 1995. Structures of genomic and complementary DNAs coding for Pleurotus ostreatus manganese(II) peroxidase. Biochim. Biophys. Acta 1251:205-209. - PubMed
-
- Cai, D., and M. Tien. 1993. Lignin-degrading peroxidases of Phanerochaete chrysosporium. J. Biotechnol. 30:79-90. - PubMed
-
- Christlet, T. H., M. Biswas, and K. Veluraja. 1999. A database analysis of potential glycosylating Asn-X-Ser/Thr consensus sequences. Acta Crystallogr. D Biol. Crystallogr. 55:1414-1420. - PubMed
-
- Cohen, R., L. Persky, and Y. Hadar. 2002. Biotechnological applications and potential of wood-degrading mushrooms of the genus Pleurotus. Appl. Microbiol. Biotechnol. 58:582-594. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

