Phosphoproteome analysis of Drosophila melanogaster embryos
- PMID: 18327897
- PMCID: PMC3063950
- DOI: 10.1021/pr700696a
Phosphoproteome analysis of Drosophila melanogaster embryos
Abstract
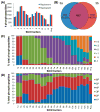
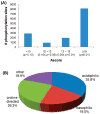
Protein phosphorylation is a key regulatory event in most cellular processes and development. Mass spectrometry-based proteomics provides a framework for the large-scale identification and characterization of phosphorylation sites. Here, we used a well-established phosphopeptide enrichment and identification strategy including the combination of strong cation exchange chromatography, immobilized metal affinity chromatography, and high-accuracy mass spectrometry instrumentation to study phosphorylation in developing Drosophila embryos. In total, 13,720 different phosphorylation sites were discovered from 2702 proteins with an estimated false-discovery rate (FDR) of 0.63% at the peptide level. Because of the large size of the data set, both novel and known phosphorylation motifs were extracted using the Motif-X algorithm, including those representative of potential ordered phosphorylation events.
Figures



References
-
- Celniker SE, Rubin GM. The Drosophila melanogaster genome. Ann Rev Genomics Hum Genet. 2003;4(1):89–117. - PubMed
-
- Adams MD. The genome sequence of Drosophila melanogaster. Science. 2000;218:5–2195. - PubMed
-
- Peter Bangs KW. Regulation and execution of apoptosis during Drosophila development. Dev Dyn. 2000;218(1):68–79. - PubMed
-
- Vidal M, Cagan RL. Drosophila models for cancer research. Curr Opin Genet Dev. 2006;16(1):10–16. - PubMed
-
- Hay BA, Guo M. Caspase-dependent cell death in Drosophila. Ann Rev Cell Dev Biol. 2006;22(1):623–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

