Rose spring dwarf-associated virus has RNA structural and gene-expression features like those of Barley yellow dwarf virus
- PMID: 18329064
- PMCID: PMC4324725
- DOI: 10.1016/j.virol.2008.01.035
Rose spring dwarf-associated virus has RNA structural and gene-expression features like those of Barley yellow dwarf virus
Abstract
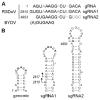
We determined the complete nucleotide sequence of the Rose spring dwarf-associated virus (RSDaV) genomic RNA (GenBank accession no. EU024678) and compared its predicted RNA structural characteristics affecting gene expression. A cDNA library was derived from RSDaV double-stranded RNAs (dsRNAs) purified from infected tissue. Nucleotide sequence analysis of the cloned cDNAs, plus for clones generated by 5'- and 3'-RACE showed the RSDaV genomic RNA to be 5808 nucleotides. The genomic RNA contains five major open reading frames (ORFs), and three small ORFs in the 3'-terminal 800 nucleotides, typical for viruses of genus Luteovirus in the family Luteoviridae. Northern blot hybridization analysis revealed the genomic RNA and two prominent subgenomic RNAs of approximately 3 kb and 1 kb. Putative 5' ends of the sgRNAs were predicted by identification of conserved sequences and secondary structures which resembled the Barley yellow dwarf virus (BYDV) genomic RNA 5' end and subgenomic RNA promoter sequences. Secondary structures of the BYDV-like ribosomal frameshift elements and cap-independent translation elements, including long-distance base pairing spanning four kb were identified. These contain similarities but also informative differences with the BYDV structures, including a strikingly different structure predicted for the 3' cap-independent translation element. These analyses of the RSDaV genomic RNA show more complexity for the RNA structural elements for members of the Luteoviridae.
Figures





Similar articles
-
Cis- and trans-regulation of luteovirus gene expression by the 3' end of the viral genome.Virus Res. 2015 Aug 3;206:37-45. doi: 10.1016/j.virusres.2015.03.009. Epub 2015 Apr 6. Virus Res. 2015. PMID: 25858272 Free PMC article. Review.
-
Molecular characterization of rose spring dwarf-associated virus isolated from China rose (Rosa chinensis Jacq.) in China.Arch Virol. 2021 Jul;166(7):2059-2062. doi: 10.1007/s00705-021-05064-4. Epub 2021 May 13. Arch Virol. 2021. PMID: 33983504
-
Nucleotide sequence shows that Bean leafroll virus has a Luteovirus-like genome organization.J Gen Virol. 2002 Jul;83(Pt 7):1791-1798. doi: 10.1099/0022-1317-83-7-1791. J Gen Virol. 2002. PMID: 12075101
-
The 3' untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element.J Virol. 2004 May;78(9):4655-64. doi: 10.1128/jvi.78.9.4655-4664.2004. J Virol. 2004. PMID: 15078948 Free PMC article.
-
Epidemiology and integrated management of persistently transmitted aphid-borne viruses of legume and cereal crops in West Asia and North Africa.Virus Res. 2009 May;141(2):209-18. doi: 10.1016/j.virusres.2008.12.007. Epub 2009 Jan 17. Virus Res. 2009. PMID: 19152820 Review.
Cited by
-
Cis- and trans-regulation of luteovirus gene expression by the 3' end of the viral genome.Virus Res. 2015 Aug 3;206:37-45. doi: 10.1016/j.virusres.2015.03.009. Epub 2015 Apr 6. Virus Res. 2015. PMID: 25858272 Free PMC article. Review.
-
Noncoding RNAs of Plant Viruses and Viroids: Sponges of Host Translation and RNA Interference Machinery.Mol Plant Microbe Interact. 2016 Mar;29(3):156-64. doi: 10.1094/MPMI-10-15-0226-FI. Epub 2016 Feb 22. Mol Plant Microbe Interact. 2016. PMID: 26900786 Free PMC article. Review.
-
Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use.Nucleic Acids Res. 2016 Sep 6;44(15):7007-78. doi: 10.1093/nar/gkw530. Epub 2016 Jul 19. Nucleic Acids Res. 2016. PMID: 27436286 Free PMC article. Review.
-
Characterization of a new apple luteovirus identified by high-throughput sequencing.Virol J. 2018 May 15;15(1):85. doi: 10.1186/s12985-018-0998-3. Virol J. 2018. PMID: 29764461 Free PMC article.
-
Translation of Plant RNA Viruses.Viruses. 2021 Dec 13;13(12):2499. doi: 10.3390/v13122499. Viruses. 2021. PMID: 34960768 Free PMC article. Review.
References
-
- Brierley I. Ribosomal frameshifting on viral RNAs. J Gen Virol. 1995;76:1885–1892. - PubMed
-
- Brierley I, Pennell S. The Ribosome. Cold Spring Harbor Symposia on Quantitative Biology, LXVI. Vol. 66. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 2001. Structure and function of the stimulatory RNAs involved in programmed eukaryotic –1 ribosomal frameshifting; pp. 233–248. - PubMed
-
- D’Arcy CJ, Domier LL. Family Luteoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press; 2005. pp. 343–352.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

