Evidence for a transposition event in a second NITR gene cluster in zebrafish
- PMID: 18330557
- PMCID: PMC2911966
- DOI: 10.1007/s00251-008-0285-3
Evidence for a transposition event in a second NITR gene cluster in zebrafish
Abstract
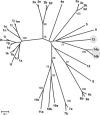
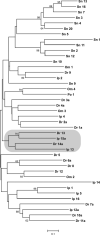
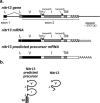
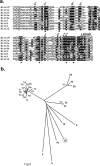
Novel immune-type receptors (NITRs) are immunoglobulin-variable (V) domain-containing cell surface proteins that possess characteristic activating/inhibitory signaling motifs and are expressed in hematopoietic cells. NITRs are encoded by multigene families and have been identified in bony fish species. A single gene cluster, which encodes 36 NITRs that can be classified into 12 families, has been mapped to zebrafish chromosome 7. We report herein the presence of a second NITR gene cluster on zebrafish chromosome 14, which is comprised of three genes (nitr13, nitr14a, and nitr14b) representing two additional NITR gene families. Phylogenetic analyses indicate that the V domains encoded by the nitr13 and nitr14 genes are more similar to each other than any other zebrafish NITR suggesting that these genes arose from a tandem gene duplication event. Similar analyses comparing zebrafish Nitr13 and Nitr14 to NITRs from other fish species indicate that the nitr13 and nitr14 genes are phylogenetically related to the catfish IpNITR13 and IpNITR15 genes. Sequence features of the chromosomal region encoding nitr13 suggest that this gene arose via retrotransposition.
Figures



References
-
- Evenhuis J, Bengten E, Snell C, Quiniou SM, Miller NW, Wilson M. Characterization of additional novel immune type receptors in channel catfish, Ictalurus punctatus. Immunogenetics. 2007 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

