Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus
- PMID: 18331720
- PMCID: PMC2279146
- DOI: 10.1016/j.devcel.2008.01.005
Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus
Abstract
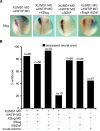
Snail family transcriptional repressors regulate epithelial mesenchymal transitions during physiological and pathological processes. A conserved SNAG repression domain present in all vertebrate Snail proteins is necessary for repressor complex assembly. Here, we identify the Ajuba family of LIM proteins as functional corepressors of the Snail family via an interaction with the SNAG domain. Ajuba LIM proteins interact with Snail in the nucleus on endogenous E-cadherin promoters and contribute to Snail-dependent repression of E-cadherin. Using Xenopus neural crest as a model of in vivo Snail- or Slug-induced EMT, we demonstrate that Ajuba LIM proteins contribute to neural crest development as Snail/Slug corepressors and are required for in vivo Snail/Slug function. Because Ajuba LIM proteins are also components of adherens junctions and contribute to their assembly or stability, their functional interaction with Snail proteins in the nucleus suggests that Ajuba LIM proteins are important regulators of epithelia dynamics communicating surface events with nuclear responses.
Figures

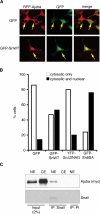
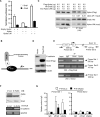
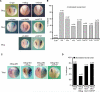
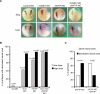

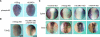
References
-
- Aybar MJ, Nieto MA, Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130(3):483–494. - PubMed
-
- Ayyanathan K, Peng H, Hou Z, Fredericks WJ, Goyal RK, Langer EM, Longmore GD, Rauscher FJ., 3rd The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic Shuttling. Cancer Res. 2007;67(19):9097–9106. - PubMed
-
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. - PubMed
-
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. - PubMed
-
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Dev Cell. 2003;4(6):827–839. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- R01 GM066815/GM/NIGMS NIH HHS/United States
- CA75315/CA/NCI NIH HHS/United States
- R01 CA092088/CA/NCI NIH HHS/United States
- R01 GM080673/GM/NIGMS NIH HHS/United States
- CA095561/CA/NCI NIH HHS/United States
- CA92088/CA/NCI NIH HHS/United States
- CA106496/CA/NCI NIH HHS/United States
- R01 CA095561/CA/NCI NIH HHS/United States
- R21 CA106496/CA/NCI NIH HHS/United States
- P30 CA010815/CA/NCI NIH HHS/United States
- GM080673/GM/NIGMS NIH HHS/United States
- CA10815/CA/NCI NIH HHS/United States
- GM66815-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

