SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus
- PMID: 18332116
- PMCID: PMC2423153
- DOI: 10.1128/MCB.02019-07
SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus
Abstract
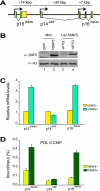
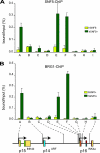
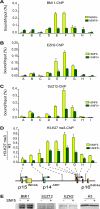
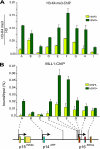
Stable silencing of the INK4b-ARF-INK4a tumor suppressor locus occurs in a variety of human cancers, including malignant rhabdoid tumors (MRTs). MRTs are extremely aggressive cancers caused by the loss of the hSNF5 subunit of the SWI/SNF chromatin-remodeling complex. We found previously that, in MRT cells, hSNF5 is required for p16(INK4a) induction, mitotic checkpoint activation, and cellular senescence. Here, we investigated how the balance between Polycomb group (PcG) silencing and SWI/SNF activation affects epigenetic control of the INK4b-ARF-INK4a locus in MRT cells. hSNF5 reexpression in MRT cells caused SWI/SNF recruitment and activation of p15(INK4b) and p16(INK4a), but not of p14(ARF). Gene activation by hSNF5 is strictly dependent on the SWI/SNF motor subunit BRG1. SWI/SNF mediates eviction of the PRC1 and PRC2 PcG silencers and extensive chromatin reprogramming. Concomitant with PcG complex removal, the mixed lineage leukemia 1 (MLL1) protein is recruited and active histone marks supplant repressive ones. Strikingly, loss of PcG complexes is accompanied by DNA methyltransferase DNMT3B dissociation and reduced DNA methylation. Thus, various chromatin states can be modulated by SWI/SNF action. Collectively, these findings emphasize the close interconnectivity and dynamics of diverse chromatin modifications in cancer and gene control.
Figures

References
-
- Agger, K., P. A. C. Cloos, J. Christensen, D. Pasini, S. Rose, J. Rappsilber, I. Issaeva, E. Canaani, A. E. Salcini, and K. Helin. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449731-734. - PubMed
-
- Bernard, D., J. F. Martinez-Leal, S. Rizzo, D. Martinez, D. Hudson, T. Visakorpi, G. Peters, A. Carnero, D. Beach, and J. Gil. 2005. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene 245543-5551. - PubMed
-
- Betz, B. L., M. W. Strobeck, D. N. Reisman, E. S. Knudsen, and B. E. Weissman. 2002. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene 215193-5203. - PubMed
-
- Biegel, J. A., J. Y. Zhou, L. B. Rorke, C. Stenstrom, L. M. Wainwright, and B. Fogelgren. 1999. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 5974-79. - PubMed
-
- Bracken, A. P., D. Kleine-Kohlbrecher, N. Dietrich, D. Pasini, G. Gargiulo, C. Beekman, K. Theilgaard-Monch, S. Minucci, B. T. Porse, J.-C. Marine, K. H. Hansen, and K. Helin. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21525-530. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
