Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation
- PMID: 18332431
- PMCID: PMC2393767
- DOI: 10.1073/pnas.0710285105
Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation
Abstract
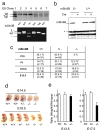
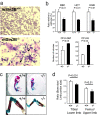
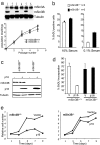
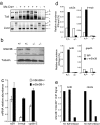
The Sin3-histone deacetylase (HDAC) corepressor complex is conserved from yeast to humans. Mammals possess two highly related Sin3 proteins, mSin3A and mSin3B, which serve as scaffolds tethering HDAC enzymatic activity, and numerous sequence-specific transcription factors to enable local chromatin regulation at specific gene targets. Despite broad overlapping expression of mSin3A and mSin3B, mSin3A is cell-essential and vital for early embryonic development. Here, genetic disruption of mSin3B reveals a very different phenotype characterized by the survival of cultured cells and lethality at late stages of embryonic development with defective differentiation of multiple lineages-phenotypes that are strikingly reminiscent of those associated with loss of retinoblastoma family members or E2F transcriptional repressors. Additionally, we observe that, whereas mSin3B(-/-) cells cycle normally under standard growth conditions, they show an impaired ability to exit the cell cycle with limiting growth factors. Correspondingly, mSin3B interacts physically with the promoters of known E2F target genes, and its deficiency is associated with derepression of these gene targets in vivo. Together, these results reveal a critical role for mSin3B in the control of cell cycle exit and terminal differentiation in mammals and establish contrasting roles for the mSin3 proteins in the growth and development of specific lineages.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. - PubMed
-
- Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2004;47:1–17. - PubMed
-
- Laherty CD, et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

