Evolution of hedgehog and hedgehog-related genes, their origin from Hog proteins in ancestral eukaryotes and discovery of a novel Hint motif
- PMID: 18334026
- PMCID: PMC2362128
- DOI: 10.1186/1471-2164-9-127
Evolution of hedgehog and hedgehog-related genes, their origin from Hog proteins in ancestral eukaryotes and discovery of a novel Hint motif
Abstract
Background: The Hedgehog (Hh) signaling pathway plays important roles in human and animal development as well as in carcinogenesis. Hh molecules have been found in both protostomes and deuterostomes, but curiously the nematode Caenorhabditis elegans lacks a bona-fide Hh. Instead a series of Hh-related proteins are found, which share the Hint/Hog domain with Hh, but have distinct N-termini.
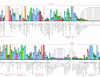
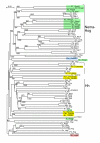
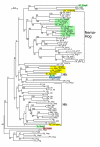
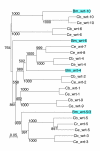
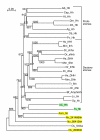
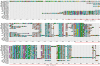
Results: We performed extensive genome searches such as the cnidarian Nematostella vectensis and several nematodes to gain further insights into Hh evolution. We found six genes in N. vectensis with a relationship to Hh: two Hh genes, one gene with a Hh N-terminal domain fused to a Willebrand factor type A domain (VWA), and three genes containing Hint/Hog domains with distinct novel N-termini. In the nematode Brugia malayi we find the same types of hh-related genes as in C. elegans. In the more distantly related Enoplea nematodes Xiphinema and Trichinella spiralis we find a bona-fide Hh. In addition, T. spiralis also has a quahog gene like C. elegans, and there are several additional hh-related genes, some of which have secreted N-terminal domains of only 15 to 25 residues. Examination of other Hh pathway components revealed that T. spiralis - like C. elegans - lacks some of these components. Extending our search to all eukaryotes, we recovered genes containing a Hog domain similar to Hh from many different groups of protists. In addition, we identified a novel Hint gene family present in many eukaryote groups that encodes a VWA domain fused to a distinct Hint domain we call Vint. Further members of a poorly characterized Hint family were also retrieved from bacteria.
Conclusion: In Cnidaria and nematodes the evolution of hh genes occurred in parallel to the evolution of other genes that contain a Hog domain but have different N-termini. The fact that Hog genes comprising a secreted N-terminus and a Hog domain are found in many protists indicates that this gene family must have arisen in very early eukaryotic evolution, and gave rise eventually to hh and hh-related genes in animals. The results indicate a hitherto unsuspected ability of Hog domain encoding genes to evolve new N-termini. In one instance in Cnidaria, the Hh N-terminal signaling domain is associated with a VWA domain and lacks a Hog domain, suggesting a modular mode of evolution also for the N-terminal domain. The Hog domain proteins, the inteins and VWA-Vint proteins are three families of Hint domain proteins that evolved in parallel in eukaryotes.
Figures







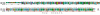


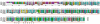


References
-
- Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development (Cambridge, England) 2006;133:3–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

