Conditional clustering of temporal expression profiles
- PMID: 18334028
- PMCID: PMC2335301
- DOI: 10.1186/1471-2105-9-147
Conditional clustering of temporal expression profiles
Abstract
Background: Many microarray experiments produce temporal profiles in different biological conditions but common cluster techniques are not able to analyze the data conditional on the biological conditions.
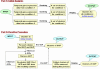
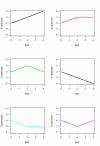
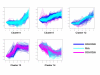
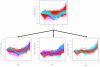
Results: This article presents a novel technique to cluster data from time course microarray experiments performed across several experimental conditions. Our algorithm uses polynomial models to describe the gene expression patterns over time, a full Bayesian approach with proper conjugate priors to make the algorithm invariant to linear transformations, and an iterative procedure to identify genes that have a common temporal expression profile across two or more experimental conditions, and genes that have a unique temporal profile in a specific condition.
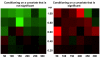
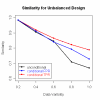
Conclusion: We use simulated data to evaluate the effectiveness of this new algorithm in finding the correct number of clusters and in identifying genes with common and unique profiles. We also use the algorithm to characterize the response of human T cells to stimulations of antigen-receptor signaling gene expression temporal profiles measured in six different biological conditions and we identify common and unique genes. These studies suggest that the methodology proposed here is useful in identifying and distinguishing uniquely stimulated genes from commonly stimulated genes in response to variable stimuli. Software for using this clustering method is available from the project home page.
Figures





References
-
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. - DOI - PMC - PubMed
-
- Golub R, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Loh ML, Coller H, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

