Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis
- PMID: 18334513
- PMCID: PMC2650625
- DOI: 10.1093/dnares/dsn002
Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis
Abstract
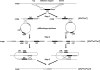
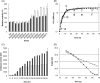
The emerging field of synthetic genomics is expected to facilitate the generation of microorganisms with the potential to achieve a sustainable society. One approach towards this goal is the reduction of microbial genomes by rationally designed deletions to create simplified cells with predictable behavior that act as a platform to build in various genetic systems for specific purposes. We report a novel Bacillus subtilis strain, MBG874, depleted of 874 kb (20%) of the genomic sequence. When compared with wild-type cells, the regulatory network of gene expression of the mutant strain is reorganized after entry into the transition state due to the synergistic effect of multiple deletions, and productivity of extracellular cellulase and protease from transformed plasmids harboring the corresponding genes is remarkably enhanced. To our knowledge, this is the first report demonstrating that genome reduction actually contributes to the creation of bacterial cells with a practical application in industry. Further systematic analysis of changes in the transcriptional regulatory network of MGB874 cells in relation to protein productivity should facilitate the generation of improved B. subtilis cells as hosts of industrial protein production.
Figures



References
-
- Ball P. Synthetic biology: designs for life. Nature. 2007;448:32–33. - PubMed
-
- Drubin D. A., Way J. C., Silver P. A. Designing biological systems. Genes Develop. 2007;21:242–254. - PubMed
-
- Forster A. C., Church G. M. Synthetic biology projects in vitro. Genome Res. 2007;17:1–6. - PubMed
-
- Posfai G., Plunkett G., III, Feher T., Frisch D., Keil G. M., Umenhoffer K., Kolisnychenko V., Stahl B., Sharma S. S., de Arruda M., et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. - PubMed
-
- Mizoguchi H., Mori H., Fujio T. Escherichia coli minimum genome factory. Biotechnol. Appl. Biochem. 2007;46:157–167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

