Calpain 3 is a modulator of the dysferlin protein complex in skeletal muscle
- PMID: 18334579
- PMCID: PMC2900895
- DOI: 10.1093/hmg/ddn081
Calpain 3 is a modulator of the dysferlin protein complex in skeletal muscle
Abstract
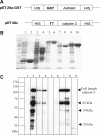
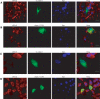
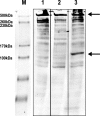
Muscular dystrophies comprise a genetically heterogeneous group of degenerative muscle disorders characterized by progressive muscle wasting and weakness. Two forms of limb-girdle muscular dystrophy, 2A and 2B, are caused by mutations in calpain 3 (CAPN3) and dysferlin (DYSF), respectively. While CAPN3 may be involved in sarcomere remodeling, DYSF is proposed to play a role in membrane repair. The coexistence of CAPN3 and AHNAK, a protein involved in subsarcolemmal cytoarchitecture and membrane repair, in the dysferlin protein complex and the presence of proteolytic cleavage fragments of AHNAK in skeletal muscle led us to investigate whether AHNAK can act as substrate for CAPN3. We here demonstrate that AHNAK is cleaved by CAPN3 and show that AHNAK is lost in cells expressing active CAPN3. Conversely, AHNAK accumulates when calpain 3 is defective in skeletal muscle of calpainopathy patients. Moreover, we demonstrate that AHNAK fragments cleaved by CAPN3 have lost their affinity for dysferlin. Thus, our findings suggest interconnectivity between both diseases by revealing a novel physiological role for CAPN3 in regulating the dysferlin protein complex.
Figures





References
-
- Dincer P., Leturcq F., Richard I., Piccolo F., Yalnizoglu D., deToma C., Akcoren Z., Broux O., Deburgrave N., Brenguier L., et al. A biochemical, genetic, and clinical survey of autosomal recessive limb girdle muscular dystrophies in Turkey. Ann. Neurol. 1997;42:222–229. - PubMed
-
- Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., et al. Mutations in the proteolytic-enzyme calpain-3 cause limb-girdle muscular-dystrophy type-2A. Cell. 1995;81:27–40. - PubMed
-
- Goll D.E., Thompson V.F., Li H., Wei W., Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

