Phenotypic mismatches reveal escape from arms-race coevolution
- PMID: 18336073
- PMCID: PMC2265764
- DOI: 10.1371/journal.pbio.0060060
Phenotypic mismatches reveal escape from arms-race coevolution
Abstract
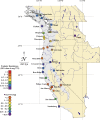
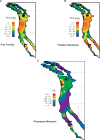
Because coevolution takes place across a broad scale of time and space, it is virtually impossible to understand its dynamics and trajectories by studying a single pair of interacting populations at one time. Comparing populations across a range of an interaction, especially for long-lived species, can provide insight into these features of coevolution by sampling across a diverse set of conditions and histories. We used measures of prey traits (tetrodotoxin toxicity in newts) and predator traits (tetrodotoxin resistance of snakes) to assess the degree of phenotypic mismatch across the range of their coevolutionary interaction. Geographic patterns of phenotypic exaggeration were similar in prey and predators, with most phenotypically elevated localities occurring along the central Oregon coast and central California. Contrary to expectations, however, these areas of elevated traits did not coincide with the most intense coevolutionary selection. Measures of functional trait mismatch revealed that over one-third of sampled localities were so mismatched that reciprocal selection could not occur given current trait distributions. Estimates of current locality-specific interaction selection gradients confirmed this interpretation. In every case of mismatch, predators were "ahead" of prey in the arms race; the converse escape of prey was never observed. The emergent pattern suggests a dynamic in which interacting species experience reciprocal selection that drives arms-race escalation of both prey and predator phenotypes at a subset of localities across the interaction. This coadaptation proceeds until the evolution of extreme phenotypes by predators, through genes of large effect, allows snakes to, at least temporarily, escape the arms race.
Conflict of interest statement
Figures


Comment in
-
Predators make (temporary) escape from coevolutionary arms race.PLoS Biol. 2008 Mar;6(3):e75. doi: 10.1371/journal.pbio.0060075. Epub 2008 Mar 11. PLoS Biol. 2008. PMID: 20076705 Free PMC article. No abstract available.
References
-
- Thompson JN. The geographic mosaic of coevolution. Chicago: University of Chicago Press; 2005. 441
-
- Thompson JN. The coevolutionary process. Chicago: University of Chicago Press; 1994. 376
-
- Thompson JN, Cunningham BM. Geographic structure and dynamics of coevolutionary selection. Nature. 2002;417:735–738. - PubMed
-
- Gomulkiewicz R, Drown DM, Dybdahl MF, Godsoe W, Nuismer SL, et al. Dos and don'ts of testing the geographic mosaic theory of coevolution. Heredity. 2007;98:249–258. - PubMed
-
- Gomulkiewicz R, Thompson JN, Holt RD, Nuismer SL, Hochberg ME. Hot spots, cold spots, and the geographic mosaic theory of coevolution. Am Nat. 2000;156:156–174. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

