Activated nuclear metabotropic glutamate receptor mGlu5 couples to nuclear Gq/11 proteins to generate inositol 1,4,5-trisphosphate-mediated nuclear Ca2+ release
- PMID: 18337251
- PMCID: PMC2376237
- DOI: 10.1074/jbc.M708551200
Activated nuclear metabotropic glutamate receptor mGlu5 couples to nuclear Gq/11 proteins to generate inositol 1,4,5-trisphosphate-mediated nuclear Ca2+ release
Abstract
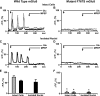
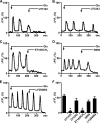
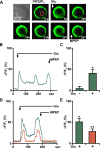
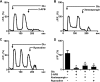
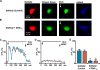
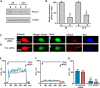
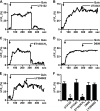
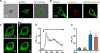
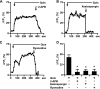
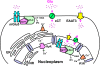
Recently we have shown that the metabotropic glutamate 5 (mGlu5) receptor can be expressed on nuclear membranes of heterologous cells or endogenously on striatal neurons where it can mediate nuclear Ca2+ changes. Here, pharmacological, optical, and genetic techniques were used to show that upon activation, nuclear mGlu5 receptors generate nuclear inositol 1,4,5-trisphosphate (IP3) in situ. Specifically, expression of an mGlu5 F767S mutant in HEK293 cells that blocks Gq/11 coupling or introduction of a dominant negative Galphaq construct in striatal neurons prevented nuclear Ca2+ changes following receptor activation. These data indicate that nuclear mGlu5 receptors couple to Gq/11 to mobilize nuclear Ca2+. Nuclear mGlu5-mediated Ca2+ responses could also be blocked by the phospholipase C (PLC) inhibitor, U73122, the phosphatidylinositol (PI) PLC inhibitor 1-O-octadecyl-2-O-methyl-sn-glycero-3-phosphorylcholine (ET-18-OCH3), or by using small interfering RNA targeted against PLCbeta1 demonstrating that PI-PLC is involved. Direct assessment of inositol phosphate production using a PIP2/IP3 "biosensor" revealed for the first time that IP3 can be generated in the nucleus following activation of nuclear mGlu5 receptors. Finally, both IP3 and ryanodine receptor blockers prevented nuclear mGlu5-mediated increases in intranuclear Ca2+. Collectively, this study shows that like plasma membrane receptors, activated nuclear mGlu5 receptors couple to Gq/11 and PLC to generate IP3-mediated release of Ca2+ from Ca2+-release channels in the nucleus. Thus the nucleus can function as an autonomous organelle independent of signals originating in the cytoplasm, and nuclear mGlu5 receptors play a dynamic role in mobilizing Ca2+ in a specific, localized fashion.
Figures

Similar articles
-
ATP causes release of intracellular Ca2+ via the phospholipase C beta/IP3 pathway in astrocytes from the dorsal spinal cord.J Neurosci. 1995 Apr;15(4):2961-71. doi: 10.1523/JNEUROSCI.15-04-02961.1995. J Neurosci. 1995. PMID: 7722640 Free PMC article.
-
Inhibition of muscarinic-stimulated polyphosphoinositide hydrolysis and Ca2+ mobilization in cat iris sphincter smooth muscle cells by cAMP-elevating agents.Cell Signal. 1997 Sep;9(6):411-21. doi: 10.1016/s0898-6568(97)00018-1. Cell Signal. 1997. PMID: 9376222
-
A role for phospholipase C activity but not ryanodine receptors in the initiation and propagation of intercellular calcium waves.J Cell Sci. 1995 Jul;108 ( Pt 7):2583-90. doi: 10.1242/jcs.108.7.2583. J Cell Sci. 1995. PMID: 7593299
-
Formation and actions of calcium-mobilizing messenger, inositol 1,4,5-trisphosphate.Am J Physiol. 1987 Feb;252(2 Pt 1):G149-57. doi: 10.1152/ajpgi.1987.252.2.G149. Am J Physiol. 1987. PMID: 3030126 Review.
-
Calcium and IP3 dynamics in cardiac myocytes: experimental and computational perspectives and approaches.Front Pharmacol. 2014 Mar 6;5:35. doi: 10.3389/fphar.2014.00035. eCollection 2014. Front Pharmacol. 2014. PMID: 24639654 Free PMC article. Review.
Cited by
-
Intracellular metabotropic glutamate receptor 5 (mGluR5) activates signaling cascades distinct from cell surface counterparts.J Biol Chem. 2009 Dec 18;284(51):35827-38. doi: 10.1074/jbc.M109.046276. J Biol Chem. 2009. PMID: 19840937 Free PMC article.
-
Cysteine (C)-x-C receptor 4 undergoes transportin 1-dependent nuclear localization and remains functional at the nucleus of metastatic prostate cancer cells.PLoS One. 2013;8(2):e57194. doi: 10.1371/journal.pone.0057194. Epub 2013 Feb 28. PLoS One. 2013. PMID: 23468933 Free PMC article.
-
The roles played by highly truncated splice variants of G protein-coupled receptors.J Mol Signal. 2012 Sep 1;7(1):13. doi: 10.1186/1750-2187-7-13. J Mol Signal. 2012. PMID: 22938630 Free PMC article.
-
Calcium signaling in synapse-to-nucleus communication.Cold Spring Harb Perspect Biol. 2011 Nov 1;3(11):a004564. doi: 10.1101/cshperspect.a004564. Cold Spring Harb Perspect Biol. 2011. PMID: 21791697 Free PMC article. Review.
-
GPCR signalling from within the cell.Br J Pharmacol. 2018 Nov;175(21):4026-4035. doi: 10.1111/bph.14023. Epub 2017 Oct 3. Br J Pharmacol. 2018. PMID: 28872669 Free PMC article. Review.
References
-
- Berridge, M. J. (2001) Novartis Found. Symp. 239 52-64 - PubMed
-
- Gerasimenko, O. V., Gerasimenko, J. V., Tepikin, A. V., and Petersen, O. H. (1995) Cell 80 439-444 - PubMed
-
- Humbert, J. P., Matter, N., Artault, J. C., Koppler, P., and Malviya, A. N. (1996) J. Biol. Chem. 271 478-485 - PubMed
-
- Alonso, M. T., Villalobos, C., Chamero, P., Alvarez, J., and Garcia-Sancho, J. (2006) Cell Calcium 40 513-525 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

