Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae
- PMID: 18337489
- PMCID: PMC2330323
- DOI: 10.1104/pp.107.114041
Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae
Abstract
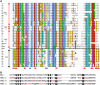
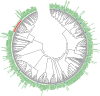
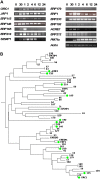
Tobacco (Nicotiana tabacum) is a member of the Solanaceae, one of the agronomically most important groups of flowering plants. We have performed an in silico analysis of 1.15 million gene-space sequence reads from the tobacco nuclear genome and report the detailed analysis of more than 2,500 tobacco transcription factors (TFs). The tobacco genome contains at least one member of each of the 64 well-characterized TF families identified in sequenced vascular plant genomes, indicating that evolution of the Solanaceae was not associated with the gain or loss of TF families. However, we found notable differences between tobacco and non-Solanaceae species in TF family size and evidence for both tobacco- and Solanaceae-specific subfamily expansions. Compared with TF families from sequenced plant genomes, tobacco has a higher proportion of ERF/AP2, C2H2 zinc finger, homeodomain, GRF, TCP, zinc finger homeodomain, BES, and STERILE APETALA (SAP) genes and novel subfamilies of BES, C2H2 zinc finger, SAP, and NAC genes. The novel NAC subfamily, termed TNACS, appears restricted to the Solanaceae, as they are absent from currently sequenced plant genomes but present in tomato (Solanum lycopersicum), pepper (Capsicum annuum), and potato (Solanum tuberosum). They constitute approximately 25% of NAC genes in tobacco. Based on our phylogenetic studies, we predict that many of the more than 50 tobacco group IX ERF genes are involved in jasmonate responses. Consistent with this, over two-thirds of group IX ERF genes tested showed increased mRNA levels following jasmonate treatment. Our data are a major resource for the Solanaceae and fill a void in studies of TF families across the plant kingdom.
Figures






References
-
- Bennetzen JL, Schrick K, Springer PS, Brown WE, Sanmiguel P (1994) Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37 565–576 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

