Toll-like receptors control autophagy
- PMID: 18337753
- PMCID: PMC2323261
- DOI: 10.1038/emboj.2008.31
Toll-like receptors control autophagy
Abstract
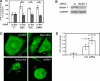
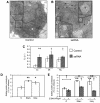
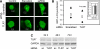
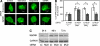
Autophagy is a newly recognized innate defense mechanism, acting as a cell-autonomous system for elimination of intracellular pathogens. The signals and signalling pathways inducing autophagy in response to pathogen invasion are presently not known. Here we show that autophagy is controlled by recognizing conserved pathogen-associated molecular patterns (PAMPs). We screened a PAMP library for effects on autophagy in RAW 264.7 macrophages and found that several prototype Toll-like receptor (TLR) ligands induced autophagy. Single-stranded RNA and TLR7 generated the most potent effects. Induction of autophagy via TLR7 depended on MyD88 expression. Stimulation of autophagy with TLR7 ligands was functional in eliminating intracellular microbes, even when the target pathogen was normally not associated with TLR7 signalling. These findings link two innate immunity defense systems, TLR signalling and autophagy, provide a potential molecular mechanism for induction of autophagy in response to pathogen invasion, and show that the newly recognized ability of TLR ligands to stimulate autophagy can be used to treat intracellular pathogens.
Figures


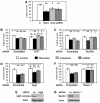
References
-
- Beutner KR, Spruance SL, Hougham AJ, Fox TL, Owens ML, Douglas JM Jr (1998) Treatment of genital warts with an immune-response modifier (imiquimod). J Am Acad Dermatol 38: 230–239 - PubMed
-
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH (2006) Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem 281: 11374–11383 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

