Investigating the regulation of brain-specific kinases 1 and 2 by phosphorylation
- PMID: 18339622
- PMCID: PMC3258900
- DOI: 10.1074/jbc.M710381200
Investigating the regulation of brain-specific kinases 1 and 2 by phosphorylation
Abstract
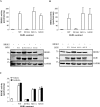
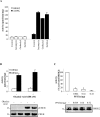
Brain-specific kinases 1 and 2 (BRSK1/2) are AMP-activated protein kinase (AMPK)-related kinases that are highly expressed in mammalian forebrain. Studies using transgenic animal models have implicated a role for these kinases in the establishment of neuronal polarity. BRSK1 and BRSK2 are activated by phosphorylation of a threonine residue in the T-loop activation segment of the kinase domain. In vitro studies have demonstrated that LKB1, an upstream kinase in the AMPK cascade, can catalyze this phosphorylation. However, to date, a detailed comparative analysis of the molecular regulation of BRSK1/2 has not been undertaken. Here we present evidence that excludes another upstream kinase in the AMPK cascade, Ca(2+)/calmodulin-dependent protein kinase kinase beta, from a role in activating BRSK1/2. We show that equivalent mutations in the ubiquitin-associated domains of the BRSK isoforms produce differential effects on the activation of BRSK1 and BRSK2. Contrary to previous reports, activation of cAMP-dependent protein kinase does not affect BRSK1 or BRSK2 activity in mammalian cells. Furthermore, stimuli that activate AMPK had no effect on BRSK1/2. Finally, we provide evidence suggesting that protein phosphatase 2C is a likely candidate for catalyzing the dephosphorylation and inactivation of BRSK1/2.
Figures
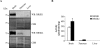
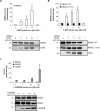
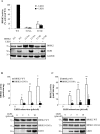
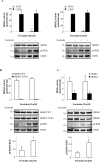
References
-
- Crump, J. G., Zhen, M., Jin, Y., and Bargmann, C. I. (2001) Neuron 29 115–129 - PubMed
-
- Inoue, E., Mochida, S., Takagi, H., Higa, S., Deguchi-Tawarada, M., Takao-Rikitsu, E., Inoue, M., Yao, I., Takeuchi, K., Kitajima, I., Setou, M., Ohtsuka, T., and Takai, Y. (2006) Neuron 50 261–275 - PubMed
-
- Kishi, M., Pan, Y. A., Crump, J. G., and Sanes, J. R. (2005) Science 307 929–932 - PubMed
-
- Hemminki, A., Markie, D., Tomlinson, I., Avizienyte, E., Roth, S., Loukola, A., Bignell, G., Warren, W., Aminoff, M., Hoglund, P., Jarvinen, H., Kristo, P., Pelin, K., Ridanpaa, M., Salovaara, R., Toro, T., Bodmer, W., Ol-schwang, S., Olsen, A. S., Stratton, M. R., de la Chapelle, A., and Aaltonen, L. A. (1998) Nature 391 184–187 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

