Paired-like homeodomain transcription factors 1 and 2 regulate follicle-stimulating hormone beta-subunit transcription through a conserved cis-element
- PMID: 18339718
- PMCID: PMC2408822
- DOI: 10.1210/en.2007-0425
Paired-like homeodomain transcription factors 1 and 2 regulate follicle-stimulating hormone beta-subunit transcription through a conserved cis-element
Abstract
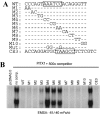
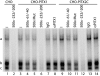
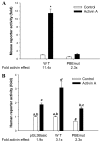
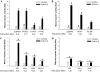
Paired-like homeodomain transcription factors (PITX) regulate the activity of pituitary hormone-encoding genes. Here, we examined mechanisms through which the family of PITX proteins control murine FSH beta-subunit (Fshb) transcription. We observed that endogenous PITX1 and PITX2 isoforms from murine LbetaT2 gonadotrope cells could bind a highly conserved proximal cis-element. Transfection of PITX1 or PITX2C in heterologous cells stimulated both murine and human Fshb/FSHB promoter-reporter activities, and in both cases, mutation of the critical cis-element abrogated these effects. In homologous LbetaT2 cells, the same mutation decreased basal reporter activity and greatly reduced activin A-stimulated transcription from murine and human promoter-reporters. Transfecting dominant-negative forms of PITX1 or PITX2C or knocking down PITX1 or -2 expression by RNA interference in LbetaT2 cells inhibited murine Fshb transcription, confirming roles for endogenous PITX proteins. Both PITX1 and PITX2C interacted with Smad3 (an effector of the activin signaling cascade in these cells) in coprecipitation experiments, and the PITX binding site mutation greatly inhibited Smad2/3/4-stimulated Fshb transcription. In summary, both PITX1 and PITX2C regulate murine and human Fshb/FSHB transcription through a conserved cis-element in the proximal promoter. Furthermore, the data indicate both common and distinct mechanisms of PITX1 and PITX2C action.
Figures


Similar articles
-
SMADs and FOXL2 synergistically regulate murine FSHbeta transcription via a conserved proximal promoter element.Mol Endocrinol. 2011 Jul;25(7):1170-83. doi: 10.1210/me.2010-0480. Epub 2011 May 26. Mol Endocrinol. 2011. PMID: 21622537 Free PMC article.
-
Activator protein-1 and smad proteins synergistically regulate human follicle-stimulating hormone beta-promoter activity.Endocrinology. 2008 Nov;149(11):5577-91. doi: 10.1210/en.2008-0220. Epub 2008 Jul 24. Endocrinology. 2008. PMID: 18653705 Free PMC article.
-
Regulation of the rat follicle-stimulating hormone beta-subunit promoter by activin.Mol Endocrinol. 2003 Mar;17(3):318-32. doi: 10.1210/me.2002-0081. Epub 2002 Dec 23. Mol Endocrinol. 2003. PMID: 12554780
-
Activin A induction of murine and ovine follicle-stimulating hormone β transcription is SMAD-dependent and TAK1 (MAP3K7)/p38 MAPK-independent in gonadotrope-like cells.Cell Signal. 2012 Aug;24(8):1632-40. doi: 10.1016/j.cellsig.2012.04.006. Epub 2012 Apr 20. Cell Signal. 2012. PMID: 22549017
-
Pitx genes in development and disease.Cell Mol Life Sci. 2021 Jun;78(11):4921-4938. doi: 10.1007/s00018-021-03833-7. Epub 2021 Apr 12. Cell Mol Life Sci. 2021. PMID: 33844046 Free PMC article. Review.
Cited by
-
Regulatory Architecture of the LβT2 Gonadotrope Cell Underlying the Response to Gonadotropin-Releasing Hormone.Front Endocrinol (Lausanne). 2018 Feb 14;9:34. doi: 10.3389/fendo.2018.00034. eCollection 2018. Front Endocrinol (Lausanne). 2018. PMID: 29487567 Free PMC article.
-
Conservation of mechanisms mediating gonadotrophin-releasing hormone 1 stimulation of human luteinizing hormone beta subunit transcription.Mol Hum Reprod. 2009 Feb;15(2):77-87. doi: 10.1093/molehr/gan079. Epub 2008 Dec 22. Mol Hum Reprod. 2009. PMID: 19106114 Free PMC article.
-
A novel role for the forkhead transcription factor FOXL2 in activin A-regulated follicle-stimulating hormone beta subunit transcription.Mol Endocrinol. 2009 Jul;23(7):1001-13. doi: 10.1210/me.2008-0324. Epub 2009 Mar 26. Mol Endocrinol. 2009. PMID: 19324968 Free PMC article.
-
Novel forms of Paired-like homeodomain transcription factor 2 (PITX2): generation by alternative translation initiation and mRNA splicing.BMC Mol Biol. 2008 Mar 28;9:31. doi: 10.1186/1471-2199-9-31. BMC Mol Biol. 2008. PMID: 18373856 Free PMC article.
-
Impaired fertility and FSH synthesis in gonadotrope-specific Foxl2 knockout mice.Mol Endocrinol. 2013 Mar;27(3):407-21. doi: 10.1210/me.2012-1286. Epub 2013 Jan 22. Mol Endocrinol. 2013. PMID: 23340250 Free PMC article.
References
-
- Brown P, McNeilly AS 1999 Transcriptional regulation of pituitary gonadotrophin subunit genes. Rev Reprod 4:117–124 - PubMed
-
- Alarid ET, Windle JJ, Whyte DB, Mellon PL 1996 Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development 122:3319–3329 - PubMed
-
- Pernasetti F, Vasilyev VV, Rosenberg SB, Bailey JS, Huang HJ, Miller WL, Mellon PL 2001 Cell-specific transcriptional regulation of follicle-stimulating hormone-β by activin and gonadotropin-releasing hormone in the LβT2 pituitary gonadotrope cell model. Endocrinology 142:2284–2295 - PubMed
-
- Gregory SJ, Lacza CT, Detz AA, Xu S, Petrillo LA, Kaiser UB 2005 Synergy between activin A and gonadotropin-releasing hormone in transcriptional activation of the rat follicle-stimulating hormone-β gene. Mol Endocrinol 19:237–254 - PubMed
-
- Bernard DJ 2004 Both SMAD2 and SMAD3 mediate activin-stimulated expression of the follicle-stimulating hormone β-subunit in mouse gonadotrope cells. Mol Endocrinol 18:606–623 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

