Mono- and trivalent ions around DNA: a small-angle scattering study of competition and interactions
- PMID: 18339743
- PMCID: PMC2426638
- DOI: 10.1529/biophysj.107.123174
Mono- and trivalent ions around DNA: a small-angle scattering study of competition and interactions
Abstract
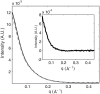
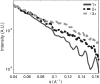
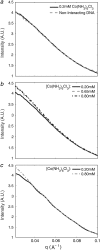
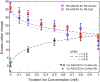
The presence of small numbers of multivalent ions in DNA-containing solutions results in strong attractive forces between DNA strands. Despite the biological importance of this interaction, e.g., DNA condensation, its physical origin remains elusive. We carried out a series of experiments to probe interactions between short DNA strands as small numbers of trivalent ions are included in a solution containing DNA and monovalent ions. Using resonant (anomalous) and nonresonant small angle x-ray scattering, we coordinated measurements of the number and distribution of each ion species around the DNA with the onset of attractive forces between DNA strands. DNA-DNA interactions occur as the number of trivalent ions increases. Surprisingly good agreement is found between data and size-corrected numerical Poisson-Boltzmann predictions of ion competition for non- and weakly interacting DNAs. We also obtained an estimate for the minimum number of trivalent ions needed to initiate DNA-DNA attraction.
Figures

References
-
- Manning, G. S. 1969. Limiting laws and counterion condensation in polyelectrolyte solutions. 1. Colligative properties. J. Chem. Phys. 51:924. - PubMed
-
- Lebret, M., and B. H. Zimm. 1984. Distribution of counterions around a cylindrical polyelectrolyte and Manning condensation theory. Biopolymers. 23:287–312. - PubMed
-
- Gueron, M., and G. Weisbuch. 1980. Poly-electrolyte theory. 1. Counterion accumulation, site-binding, and their insensitivity to poly-electrolyte shape in solutions containing finite salt concentrations. Biopolymers. 19:353–382.
-
- Nguyen, T. T., I. Rouzina, and B. I. Shklovskii. 2000. Reentrant condensation of DNA induced by multivalent counterions. J. Chem. Phys. 112:2562–2568.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

