Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export
- PMID: 18344340
- PMCID: PMC2394876
- DOI: 10.1128/AEM.02591-07
Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export
Abstract
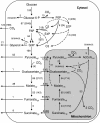
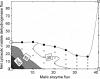
Malic acid is a potential biomass-derivable "building block" for chemical synthesis. Since wild-type Saccharomyces cerevisiae strains produce only low levels of malate, metabolic engineering is required to achieve efficient malate production with this yeast. A promising pathway for malate production from glucose proceeds via carboxylation of pyruvate, followed by reduction of oxaloacetate to malate. This redox- and ATP-neutral, CO(2)-fixing pathway has a theoretical maximum yield of 2 mol malate (mol glucose)(-1). A previously engineered glucose-tolerant, C(2)-independent pyruvate decarboxylase-negative S. cerevisiae strain was used as the platform to evaluate the impact of individual and combined introduction of three genetic modifications: (i) overexpression of the native pyruvate carboxylase encoded by PYC2, (ii) high-level expression of an allele of the MDH3 gene, of which the encoded malate dehydrogenase was retargeted to the cytosol by deletion of the C-terminal peroxisomal targeting sequence, and (iii) functional expression of the Schizosaccharomyces pombe malate transporter gene SpMAE1. While single or double modifications improved malate production, the highest malate yields and titers were obtained with the simultaneous introduction of all three modifications. In glucose-grown batch cultures, the resulting engineered strain produced malate at titers of up to 59 g liter(-1) at a malate yield of 0.42 mol (mol glucose)(-1). Metabolic flux analysis showed that metabolite labeling patterns observed upon nuclear magnetic resonance analyses of cultures grown on (13)C-labeled glucose were consistent with the envisaged nonoxidative, fermentative pathway for malate production. The engineered strains still produced substantial amounts of pyruvate, indicating that the pathway efficiency can be further improved.
Figures




Similar articles
-
Anaplerotic role for cytosolic malic enzyme in engineered Saccharomyces cerevisiae strains.Appl Environ Microbiol. 2011 Feb;77(3):732-8. doi: 10.1128/AEM.02132-10. Epub 2010 Dec 3. Appl Environ Microbiol. 2011. PMID: 21131518 Free PMC article.
-
[Construction and fermentation control of reductive TCA pathway for malic acid production in Saccharomyces cerevisiae].Sheng Wu Gong Cheng Xue Bao. 2013 Oct;29(10):1484-93. Sheng Wu Gong Cheng Xue Bao. 2013. PMID: 24432663 Chinese.
-
Metabolic engineering of Torulopsis glabrata for malate production.Metab Eng. 2013 Sep;19:10-6. doi: 10.1016/j.ymben.2013.05.002. Epub 2013 May 23. Metab Eng. 2013. PMID: 23707987
-
Malo-ethanolic fermentation in Saccharomyces and Schizosaccharomyces.Curr Genet. 2003 Sep;43(6):379-91. doi: 10.1007/s00294-003-0411-6. Epub 2003 Jun 12. Curr Genet. 2003. PMID: 12802505 Review.
-
Carboxylation and anaplerosis in neurons and glia.Mol Neurobiol. 2000 Aug-Dec;22(1-3):21-40. doi: 10.1385/MN:22:1-3:021. Mol Neurobiol. 2000. PMID: 11414279 Review.
Cited by
-
Computational approaches for microalgal biofuel optimization: a review.Biomed Res Int. 2014;2014:649453. doi: 10.1155/2014/649453. Epub 2014 Sep 21. Biomed Res Int. 2014. PMID: 25309916 Free PMC article. Review.
-
Efficient itaconic acid production from glycerol with Ustilago vetiveriae TZ1.Biotechnol Biofuels. 2017 May 19;10:131. doi: 10.1186/s13068-017-0809-x. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28533815 Free PMC article.
-
Dewar Heterocycles as Versatile Monomers for Ring-Opening Metathesis Polymerization.ACS Macro Lett. 2020 May 19;9(5):731-735. doi: 10.1021/acsmacrolett.0c00227. Epub 2020 May 1. ACS Macro Lett. 2020. PMID: 34306822 Free PMC article.
-
Investigation of malic acid production in Aspergillus oryzae under nitrogen starvation conditions.Appl Environ Microbiol. 2013 Oct;79(19):6050-8. doi: 10.1128/AEM.01445-13. Epub 2013 Jul 26. Appl Environ Microbiol. 2013. PMID: 23892740 Free PMC article.
-
Adaptation and transcriptome analysis of Aureobasidium pullulans in corncob hydrolysate for increased inhibitor tolerance to malic acid production.PLoS One. 2015 Mar 20;10(3):e0121416. doi: 10.1371/journal.pone.0121416. eCollection 2015. PLoS One. 2015. PMID: 25793624 Free PMC article.
References
-
- Abe, S., A. Furuya, T. Saito, and K. Takayama. November 1962. Method of producing L-malic acid by fermentation. U.S. patent 3,063,910.
-
- Aliverdieva, D. A., D. V. Mamaev, D. I. Bondarenko, and K. F. Sholtz. 2006. Properties of yeast Saccharomyces cerevisiae plasma membrane dicarboxylate transporter. Biochemistry (Moscow) 71:1161-1169. - PubMed
-
- Bakker, B. M., K. M. Overkamp, A. J. A. van Maris, P. Kotter, M. A. H. Luttik, J. P. van Dijken, and J. T. Pronk. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:15-37. - PubMed
-
- Battat, E., Y. Peleg, A. Bercovitz, J. S. Rokem, and I. Goldberg. 1991. Optimization of l-malic acid production by Aspergillus flavus in a stirred fermentor. Biotechnol. Bioeng. 37:1108-1116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

