Genomic insights into Mn(II) oxidation by the marine alphaproteobacterium Aurantimonas sp. strain SI85-9A1
- PMID: 18344346
- PMCID: PMC2394881
- DOI: 10.1128/AEM.01656-07
Genomic insights into Mn(II) oxidation by the marine alphaproteobacterium Aurantimonas sp. strain SI85-9A1
Abstract
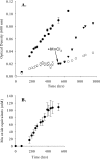
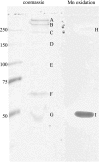
Microbial Mn(II) oxidation has important biogeochemical consequences in marine, freshwater, and terrestrial environments, but many aspects of the physiology and biochemistry of this process remain obscure. Here, we report genomic insights into Mn(II) oxidation by the marine alphaproteobacterium Aurantimonas sp. strain SI85-9A1, isolated from the oxic/anoxic interface of a stratified fjord. The SI85-9A1 genome harbors the genetic potential for metabolic versatility, with genes for organoheterotrophy, methylotrophy, oxidation of sulfur and carbon monoxide, the ability to grow over a wide range of O(2) concentrations (including microaerobic conditions), and the complete Calvin cycle for carbon fixation. Although no growth could be detected under autotrophic conditions with Mn(II) as the sole electron donor, cultures of SI85-9A1 grown on glycerol are dramatically stimulated by addition of Mn(II), suggesting an energetic benefit from Mn(II) oxidation. A putative Mn(II) oxidase is encoded by duplicated multicopper oxidase genes that have a complex evolutionary history including multiple gene duplication, loss, and ancient horizontal transfer events. The Mn(II) oxidase was most abundant in the extracellular fraction, where it cooccurs with a putative hemolysin-type Ca(2+)-binding peroxidase. Regulatory elements governing the cellular response to Fe and Mn concentration were identified, and 39 targets of these regulators were detected. The putative Mn(II) oxidase genes were not among the predicted targets, indicating that regulation of Mn(II) oxidation is controlled by other factors yet to be identified. Overall, our results provide novel insights into the physiology and biochemistry of Mn(II) oxidation and reveal a genome specialized for life at the oxic/anoxic interface.
Figures



References
-
- Al-Maghrebi, M., I. Fridovich, and L. Benov. 2002. Manganese supplementation relieves the phenotypic deficits seen in superoxide-dismutase-null Escherichia coli. Arch. Biochem. Biophys. 402:104-109. - PubMed
-
- Arp, D. J., P. S. G. Chain, and M. G. Klotz. 2007. The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu. Rev. Microbiol. 61:503-528. - PubMed
-
- Ashida, H., A. Danchin, and A. Yokota. 2005. Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism? Res. Microbiol. 156:611-618. - PubMed
-
- Bargar, J. R., B. M. Tebo, U. Bergmann, S. M. Webb, P. Glatzel, V. Q. Chiu, and M. Villalobos. 2005. Biotic and abiotic products of Mn(II) oxidation by spores of the marine Bacillus sp. strain SG-1. Am. Mineral. 90:143-154.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

