Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development
- PMID: 18344418
- PMCID: PMC2492646
- DOI: 10.1104/pp.108.116863
Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development
Abstract
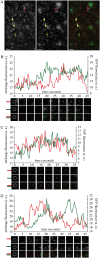
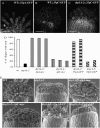
Members of the Arabidopsis (Arabidopsis thaliana) DYNAMIN-RELATED PROTEIN1 (DRP1) family are required for cytokinesis and cell expansion. Two isoforms, DRP1A and DRP1C, are required for plasma membrane maintenance during stigmatic papillae expansion and pollen development, respectively. It is unknown whether the DRP1s function interchangeably or if they have distinct roles during cell division and expansion. DRP1C was previously shown to form dynamic foci in the cell cortex, which colocalize with part of the clathrin endocytic machinery in plants. DRP1A localizes to the plasma membrane, but its cortical organization and dynamics have not been determined. Using dual color labeling with live cell imaging techniques, we showed that DRP1A also forms discreet dynamic foci in the epidermal cell cortex. Although the foci overlap with those formed by DRP1C and clathrin light chain, there are clear differences in behavior and response to pharmacological inhibitors between DRP1A and DRP1C foci. Possible functional or regulatory differences between DRP1A and DRP1C were supported by the failure of DRP1C to functionally compensate for the absence of DRP1A. Our studies indicated that the DRP1 isoforms function or are regulated differently during cell expansion.
Figures


References
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 - PubMed
-
- Banbury DN, Oakley JD, Sessions RB, Banting G (2003) Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J Biol Chem 278 12022–12028 - PubMed
-
- Baskin TI, Wilson JE, Cork A, Williamson RE (1994) Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol 35 935–942 - PubMed
-
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 - PubMed
-
- Conner SD, Schmid SL (2003) Regulated portals of entry into the cell. Nature 422 37–44 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

