NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
- PMID: 18344977
- PMCID: PMC2659626
- DOI: 10.1038/nchembio.76
NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Erratum in
- Nat Chem Biol. 2008 Jun;4(6):379
Abstract
Our understanding of how steroid hormones regulate physiological functions has been significantly advanced by structural biology approaches. However, progress has been hampered by misfolding of the ligand binding domains in heterologous expression systems and by conformational flexibility that interferes with crystallization. Here, we show that protein folding problems that are common to steroid hormone receptors are circumvented by mutations that stabilize well-characterized conformations of the receptor. We use this approach to present the structure of an apo steroid receptor that reveals a ligand-accessible channel allowing soaking of preformed crystals. Furthermore, crystallization of different pharmacological classes of compounds allowed us to define the structural basis of NFkappaB-selective signaling through the estrogen receptor, thus revealing a unique conformation of the receptor that allows selective suppression of inflammatory gene expression. The ability to crystallize many receptor-ligand complexes with distinct pharmacophores allows one to define structural features of signaling specificity that would not be apparent in a single structure.
Figures



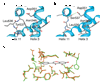
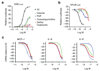
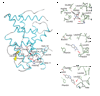
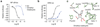
Comment in
-
Fast-tracking steroid receptor crystallization.Nat Chem Biol. 2008 Apr;4(4):226-7. doi: 10.1038/nchembio0408-226. Nat Chem Biol. 2008. PMID: 18347588 No abstract available.
References
-
- Schulman IG, Heyman RA. The flip side: Identifying small molecule regulators of nuclear receptors. Chem Biol. 2004;11:639–646. - PubMed
-
- Steffan RJ, et al. Synthesis and activity of substituted 4-(indazol-3-yl)phenols as pathway-selective estrogen receptor ligands useful in the treatment of rheumatoid arthritis. J Med Chem. 2004;47:6435–6438. - PubMed
-
- Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- PubChem-Substance/47213210
- PubChem-Substance/47213211
- PubChem-Substance/47213212
- PubChem-Substance/47213213
- PubChem-Substance/47213214
- PubChem-Substance/47213215
- PubChem-Substance/47213216
- PubChem-Substance/47213217
- PubChem-Substance/47213218
- PubChem-Substance/47213219
- PubChem-Substance/47213220
- PubChem-Substance/47213221
- PubChem-Substance/47213222
- PubChem-Substance/47213223
- PubChem-Substance/47213224
- PubChem-Substance/47213225
- PubChem-Substance/47213226
- PubChem-Substance/47213227
- PubChem-Substance/47213228
- PubChem-Substance/47213229
- PubChem-Substance/47213230
- PubChem-Substance/47213231
- PubChem-Substance/47213232
- PubChem-Substance/47213233
- PubChem-Substance/47213234
- PubChem-Substance/47213235
- PubChem-Substance/47213236
- PubChem-Substance/47213237
- PubChem-Substance/47213238
- PubChem-Substance/47213239
- PubChem-Substance/47213240
- PubChem-Substance/47213241
- PubChem-Substance/47213242
- PubChem-Substance/47213243
- PubChem-Substance/47213244
- PubChem-Substance/47213245
- PubChem-Substance/47213246
- PubChem-Substance/47213247
- PubChem-Substance/47213248
- PubChem-Substance/47213249
- PubChem-Substance/47213250
- PubChem-Substance/47213251
- PubChem-Substance/47213252
- PubChem-Substance/47213253
- PubChem-Substance/47213254
- PubChem-Substance/47213255
- PubChem-Substance/47213256
Grants and funding
- 5R01 CA18119/CA/NCI NIH HHS/United States
- R01 CA037799/CA/NCI NIH HHS/United States
- R37 DK015556/DK/NIDDK NIH HHS/United States
- R01 HL61432/HL/NHLBI NIH HHS/United States
- 5R37 DK15556/DK/NIDDK NIH HHS/United States
- R01 HL061432/HL/NHLBI NIH HHS/United States
- R01 CA018119/CA/NCI NIH HHS/United States
- R01 DK077085/DK/NIDDK NIH HHS/United States
- R33 CA132022/CA/NCI NIH HHS/United States
- 1R21 NS056998-01/NS/NINDS NIH HHS/United States
- 5R01 CA89489/CA/NCI NIH HHS/United States
- R21 NS056998/NS/NINDS NIH HHS/United States
- R01 DK015556/DK/NIDDK NIH HHS/United States
- R01 CA089489/CA/NCI NIH HHS/United States
- R01 CA37799/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

