Hypermethylated MAL gene - a silent marker of early colon tumorigenesis
- PMID: 18346269
- PMCID: PMC2292685
- DOI: 10.1186/1479-5876-6-13
Hypermethylated MAL gene - a silent marker of early colon tumorigenesis
Abstract
Background: Tumor-derived aberrantly methylated DNA might serve as diagnostic biomarkers for cancer, but so far, few such markers have been identified. The aim of the present study was to investigate the potential of the MAL (T-cell differentiation protein) gene as an early epigenetic diagnostic marker for colorectal tumors.
Methods: Using methylation-specific polymerase chain reaction (MSP) the promoter methylation status of MAL was analyzed in 218 samples, including normal mucosa (n = 44), colorectal adenomas (n = 63), carcinomas (n = 65), and various cancer cell lines (n = 46). Direct bisulphite sequencing was performed to confirm the MSP results. MAL gene expression was investigated with real time quantitative analyses before and after epigenetic drug treatment. Immunohistochemical analysis of MAL was done using normal colon mucosa samples (n = 5) and a tissue microarray with 292 colorectal tumors.
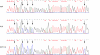
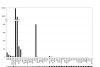
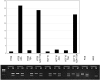
Results: Bisulphite sequencing revealed that the methylation was unequally distributed within the MAL promoter and by MSP analysis a region close to the transcription start point was shown to be hypermethylated in the majority of colorectal carcinomas (49/61, 80%) as well as in adenomas (45/63, 71%). In contrast, only a minority of the normal mucosa samples displayed hypermethylation (1/23, 4%). The hypermethylation of MAL was significantly associated with reduced or lost gene expression in in vitro models. Furthermore, removal of the methylation re-induced gene expression in colon cancer cell lines. Finally, MAL protein was expressed in epithelial cells of normal colon mucosa, but not in the malignant cells of the same type.
Conclusion: Promoter hypermethylation of MAL was present in the vast majority of benign and malignant colorectal tumors, and only rarely in normal mucosa, which makes it suitable as a diagnostic marker for early colorectal tumorigenesis.
Figures



References
-
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. - PubMed
-
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. - DOI - PMC - PubMed
-
- Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. - PubMed
-
- Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Gonzalez S, Tarafa G, Sidransky D, Meltzer SJ, Baylin SB, Herman JG. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

