The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor
- PMID: 18347045
- PMCID: PMC2423098
- DOI: 10.1128/IAI.00102-08
The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor
Abstract
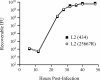
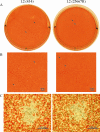
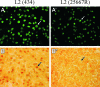
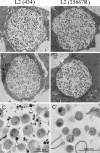
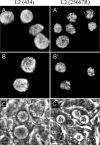
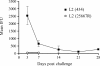
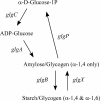
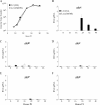
Chlamydia trachomatis possesses a cryptic 7.5-kb plasmid of unknown function. Here, we describe a comprehensive molecular and biological characterization of the naturally occurring plasmidless human C. trachomatis strain L2(25667R). We found that despite minimal chromosomal polymorphisms, the LGV strain L2(25667R) was indistinguishable from plasmid-positive strain L2(434) with regard to its in vitro infectivity characteristics such as growth kinetics, plaquing efficiency, and plaque size. The only in vitro phenotypic differences between L2(434) and L2(25667R) were the accumulation of glycogen granules in the inclusion matrix and the lack of the typical intrainclusion Brownian-like movement characteristic of C. trachomatis strains. Conversely, we observed a marked difference between the two strains in their abilities to colonize and infect the female mouse genital tract. The 50% infective dose of plasmidless strain L2(25667R) was 400-fold greater (4 x 10(6) inclusion-forming units [IFU]) than that of plasmid-bearing strain L2(434) (1 x 10(4) IFU). Transcriptome analysis of the two strains demonstrated a decrease in the transcript levels of a subset of chromosomal genes for strain L2(25667R). Among those genes was glgA, encoding glycogen synthase, a finding consistent with the failure of L2(25667R) to accumulate glycogen granules. These findings support a primary role for the plasmid in in vivo infectivity and suggest that virulence is controlled, at least in part, by the plasmid's ability to regulate the expression of chromosomal genes. Our findings have important implications in understanding a role for the plasmid in the pathogenesis of human infection and disease.
Figures

References
-
- Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2951-953. - PubMed
-
- An, Q., G. Radcliffe, R. Vassallo, D. Buxton, W. J. O'Brien, D. A. Pelletier, W. G. Weisburg, J. D. Klinger, and D. M. Olive. 1992. Infection with a plasmid-free variant chlamydia related to Chlamydia trachomatis identified by using multiple assays for nucleic acid detection. J. Clin. Microbiol. 302814-2821. - PMC - PubMed
-
- Azuma, Y., H. Hirakawa, A. Yamashita, Y. Cai, M. A. Rahman, H. Suzuki, S. Mitaku, H. Toh, S. Goto, T. Murakami, K. Sugi, H. Hayashi, H. Fukushi, M. Hattori, S. Kuhara, and M. Shirai. 2006. Genome sequence of the cat pathogen, Chlamydophilia felis. DNA Res. 1315-23. - PubMed
-
- Bai, G., E. Smith, A. Golubov, J. Pata, and K. A. McDonough. 2007. Differential gene regulation in Yersinia pestis pseudotuberculosis: effects of hypoxia and potential role of a plasmid regulator. Adv. Exp. Med. Biol. 603131-144. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

