Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B
- PMID: 18347089
- PMCID: PMC2394721
- DOI: 10.1101/gad.1643908
Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B
Abstract
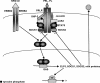
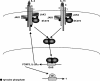
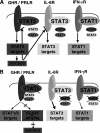
Transcription factors from the family of Signal Transducers and Activators of Transcription (STAT) are activated by numerous cytokines. Two members of this family, STAT5A and STAT5B (collectively called STAT5), have gained prominence in that they are activated by a wide variety of cytokines such as interleukins, erythropoietin, growth hormone, and prolactin. Furthermore, constitutive STAT5 activation is observed in the majority of leukemias and many solid tumors. Inactivation studies in mice as well as human mutations have provided insight into many of STAT5's functions. Disruption of cytokine signaling through STAT5 results in a variety of cell-specific effects, ranging from a defective immune system and impaired erythropoiesis, the complete absence of mammary development during pregnancy, to aberrant liver function. On a molecular level, STAT5 has been linked to cell specification, proliferation, differentiation, and survival. Evidence is growing that the diverse outcomes of STAT5 signaling are not only determined by the expression of specific receptors but also by the interaction of STAT5 with cofactors and the cell-specific activity of members of the SOCS family, which negatively regulate STAT function. In this review, we focus on emerging concepts and challenges in the field of Janus kinase (JAK)-STAT5 signaling. First, we discuss unique functions of STAT5 in three distinct systems: mammary epithelial cells, hepatocytes, and regulatory T cells. Second, we present an example of how STAT5 can achieve cell specificity in hepatocytes through a physical and functional interaction with the glucocorticoid receptor. Third, we focus on the relevance of STAT5 in the development and progression of leukemia. Next, we discuss lessons derived from human mutations and disease. Finally, we address an emerging issue that the interpretation of experiments from STAT5-deficient mice and cells might be compromised as these cells might reroute and reprogram cytokine signals to the "wrong" STATs and thus acquire inappropriate cues. We propose that mice with mutations in various components of the JAK-STAT signaling pathway are living laboratories, which will provide insight into the versatility of signaling hardware and the adaptability of the software.
Figures

References
-
- Bernasconi A., Marino R., Ribas A., Rossi J., Ciaccio M., Oleastro M., Ornani A., Paz R., Rivarola M.A., Zelazko M., et al. Characterization of immunodeficiency in a patient with growth hormone insensitivity secondary to a novel STAT5b gene mutation. Pediatrics. 2006;118:e1584–e1592. doi: 10.1542/peds.2005-2882. - DOI - PubMed
-
- Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., Farrar M.A. IL-2 receptor β-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007;178:280–290. - PubMed
-
- Chen Y., Wen R., Yang S., Schuman J., Zhang E.E., Yi T., Feng G.S., Wang D. Identification of Shp-2 as a Stat5A phosphatase. J. Biol. Chem. 2003;278:16520–16527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
