Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells
- PMID: 18347094
- PMCID: PMC2275428
- DOI: 10.1101/gad.1642408
Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells
Abstract
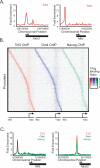
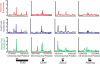
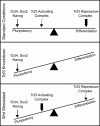
Embryonic stem (ES) cells have a unique regulatory circuitry, largely controlled by the transcription factors Oct4, Sox2, and Nanog, which generates a gene expression program necessary for pluripotency and self-renewal. How external signals connect to this regulatory circuitry to influence ES cell fate is not known. We report here that a terminal component of the canonical Wnt pathway in ES cells, the transcription factor T-cell factor-3 (Tcf3), co-occupies promoters throughout the genome in association with the pluripotency regulators Oct4 and Nanog. Thus, Tcf3 is an integral component of the core regulatory circuitry of ES cells, which includes an autoregulatory loop involving the pluripotency regulators. Both Tcf3 depletion and Wnt pathway activation cause increased expression of Oct4, Nanog, and other pluripotency factors and produce ES cells that are refractory to differentiation. Our results suggest that the Wnt pathway, through Tcf3, brings developmental signals directly to the core regulatory circuitry of ES cells to influence the balance between pluripotency and differentiation.
Figures


References
-
- Anton R., Kestler H.A., Kuhl M. β-Catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Lett. 2007;581:5247–5254. - PubMed
-
- Behrens J., von Kries J.P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. - PubMed
-
- Boiani M., Scholer H.R. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2005;6:872–884. - PubMed
-
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
