Crystal structure of a functional dimer of the PhoQ sensor domain
- PMID: 18348979
- PMCID: PMC2376233
- DOI: 10.1074/jbc.M710592200
Crystal structure of a functional dimer of the PhoQ sensor domain
Abstract
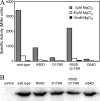
The PhoP-PhoQ two-component system is a well studied bacterial signaling system that regulates virulence and stress response. Catalytic activity of the histidine kinase sensor protein PhoQ is activated by low extracellular concentrations of divalent cations such as Mg2+, and subsequently the response regulator PhoP is activated in turn through a classic phosphotransfer pathway that is typical in such systems. The PhoQ sensor domains of enteric bacteria contain an acidic cluster of residues (EDDDDAE) that has been implicated in direct binding to divalent cations. We have determined crystal structures of the wild-type Escherichia coli PhoQ periplasmic sensor domain and of a mutant variant in which the acidic cluster was neutralized to conservative uncharged residues (QNNNNAQ). The PhoQ domain structure is similar to that of DcuS and CitA sensor domains, and this PhoQ-DcuS-CitA (PDC) sensor fold is seen to be distinct from the superficially similar PAS domain fold. Analysis of the wild-type structure reveals a dimer that allows for the formation of a salt bridge across the dimer interface between Arg-50' and Asp-179 and with nickel ions bound to aspartate residues in the acidic cluster. The physiological importance of the salt bridge to in vivo PhoQ function has been confirmed by mutagenesis. The mutant structure has an alternative, non-physiological dimeric association.
Figures
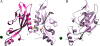


References
-
- Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000) Annu. Rev. Biochem. 69 183-215 - PubMed
-
- West, A. H., and Stock, A. M. (2001) Trends Biochem. Sci. 26 369-376 - PubMed
-
- Wolanin, P. M., Thomason, P. A., and Stock, J. B. (2002) Genome Biology http://genomebiology.com/2002/3/10/reviews/3013 - PMC - PubMed
-
- Robinson, V. L., Buckler, D. R., and Stock, A. M. (2000) Nat. Struct. Biol. 7 626-633 - PubMed
-
- Dutta, R., Qin, L., and Inouye, M. (1999) Mol. Microbiol. 34 633-640 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

