E2F1 regulates the base excision repair gene XRCC1 and promotes DNA repair
- PMID: 18348985
- PMCID: PMC2397471
- DOI: 10.1074/jbc.M710296200
E2F1 regulates the base excision repair gene XRCC1 and promotes DNA repair
Abstract
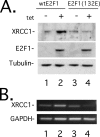
The E2F1 transcription factor activates S-phase-promoting genes, mediates apoptosis, and stimulates DNA repair through incompletely understood mechanisms. XRCC1 (x-ray repair cross-complementing group 1) protein is important for efficient single strand break/base excision repair. Although both damage and proliferative signals increase XRCC1 levels, the mechanisms regulating XRCC1 transcription remain unclear. To study these upstream mechanisms, the XRCC1 promoter was cloned into a luciferase reporter. Ectopic expression of wild-type E2F1, but not an inactive mutant E2F1(132E), activated the XRCC1 promoter-luciferase reporter, and deletion of predicted E2F1 binding sites in the promoter attenuated E2F1-induced activation. Endogenous XRCC1 expression increased in cells conditionally expressing wild-type, but not mutant E2F1, and methyl methanesulfonate-induced DNA damage stimulated XRCC1 expression in E2F1(+/+) but not E2F1(-/-) mouse embryo fibroblasts (MEFs). Additionally, E2F1(-/-) MEFs displayed attenuated DNA repair after methyl methanesulfonate-induced damage compared with E2F1(+/+) MEFs. Moreover, Chinese hamster ovary cells with mutant XRCC1 (EM9) were more sensitive to E2F1-induced apoptosis compared with Chinese hamster ovary cells with wild-type XRCC1 (AA8). These results provide new mechanistic insight into the role of the E2F pathway in maintaining genomic stability.
Figures




References
-
- Sears, R., and Nevins, J. (2002) J. Biol. Chem. 277 11617-11620 - PubMed
-
- Ginsberg, D. (2002) FEBS Lett. 529 122-125 - PubMed
-
- Leone, G., Sears, R., Huang, E., Rempel, R., Nuckolls, F., Park, C.-H., Giangrande, P., Wu, L., Saavedra, H., Field, S., Thompson, M., Yang, H., Fujiwara, Y., Greenberg, M., Orkin, S., Smith, C., and Nevins, J. (2001) Mol. Cell 8 105-113 - PubMed
-
- Trimarchi, J., and Lees, J. (2002) Nat. Rev. Mol. Cell Biol. 3 11-20 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

