EPO receptor circuits for primary erythroblast survival
- PMID: 18349318
- PMCID: PMC2396729
- DOI: 10.1182/blood-2007-10-119743
EPO receptor circuits for primary erythroblast survival
Abstract
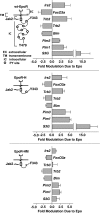
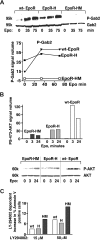
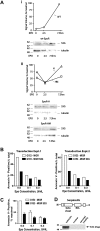
EPO functions primarily as an erythroblast survival factor, and its antiapoptotic actions have been proposed to involve predominantly PI3-kinase and BCL-X pathways. Presently, the nature of EPO-regulated survival genes has been investigated through transcriptome analyses of highly responsive, primary bone marrow erythroblasts. Two proapoptotic factors, Bim and FoxO3a, were rapidly repressed not only via the wild-type EPOR, but also by PY-deficient knocked-in EPOR alleles. In parallel, Pim1 and Pim3 kinases and Irs2 were induced. For this survival gene set, induction failed via a PY-null EPOR-HM allele, but was restored upon reconstitution of a PY343 STAT5-binding site within a related EPOR-H allele. Notably, EPOR-HM supports erythropoiesis at steady state but not during anemia, while EPOR-H exhibits near wild-type EPOR activities. EPOR-H and the wild-type EPOR (but not EPOR-HM) also markedly stimulated the expression of Trb3 pseudokinase, and intracellular serpin, Serpina-3G. For SERPINA-3G and TRB3, ectopic expression in EPO-dependent progenitors furthermore significantly inhibited apoptosis due to cytokine withdrawal. BCL-XL and BCL2 also were studied, but in highly responsive Kit(pos)CD71(high)Ter119(neg) erythroblasts, neither was EPO modulated. EPOR survival circuits therefore include the repression of Bim plus FoxO3a, and EPOR/PY343/STAT5-dependent stimulation of Pim1, Pim3, Irs2 plus Serpina-3G, and Trb3 as new antiapoptotic effectors.
Figures




References
-
- Fisher JW, Koury S, Ducey T, Mendel S. Erythropoietin production by interstitial cells of hypoxic monkey kidneys. Br J Haematol. 1996;95:27–32. - PubMed
-
- Wu H, Liu X, Jaenisch R, Lodish HF. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 1995;83:59–67. - PubMed
-
- Koury MJ, Bondurant MC. Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science. 1990;248:378–381. - PubMed
-
- Jelkmann W. Erythropoietin after a century of research: younger than ever. Eur J Haematol. 2007;78:183–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

