Velvet: algorithms for de novo short read assembly using de Bruijn graphs
- PMID: 18349386
- PMCID: PMC2336801
- DOI: 10.1101/gr.074492.107
Velvet: algorithms for de novo short read assembly using de Bruijn graphs
Abstract
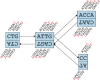
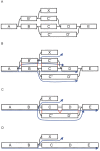
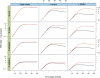
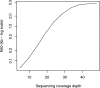
We have developed a new set of algorithms, collectively called "Velvet," to manipulate de Bruijn graphs for genomic sequence assembly. A de Bruijn graph is a compact representation based on short words (k-mers) that is ideal for high coverage, very short read (25-50 bp) data sets. Applying Velvet to very short reads and paired-ends information only, one can produce contigs of significant length, up to 50-kb N50 length in simulations of prokaryotic data and 3-kb N50 on simulated mammalian BACs. When applied to real Solexa data sets without read pairs, Velvet generated contigs of approximately 8 kb in a prokaryote and 2 kb in a mammalian BAC, in close agreement with our simulated results without read-pair information. Velvet represents a new approach to assembly that can leverage very short reads in combination with read pairs to produce useful assemblies.
Figures
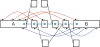
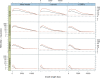
References
-
- Batzoglou S. Algorithmic challenges in mammalian genome sequence assembly. In: Dunn M., et al., editors. Encyclopedia of genomics, proteomics and bioinformatics. John Wiley and Sons; New York: 2005. Part 4.
-
- Batzoglou S., Jaffe D.B., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Jaffe D.B., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Berger B., Mesirov J.P., Lander E.S., Mesirov J.P., Lander E.S., Lander E.S. ARACHNE: A whole genome shotgun assembler. Genome Res. 2002;12:177–189. - PMC - PubMed
-
- Bentley D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. - PubMed
-
- Bokhari S.H., Sauer J.R., Sauer J.R. A parallel graph decomposition algorithm for DNA sequencing with nanopores. Bioinformatics. 2005;21:889–896. - PubMed
-
- Chaisson M., Pevzner P.A., Tang H., Pevzner P.A., Tang H., Tang H. Fragment assembly with short reads. Bioinformatics. 2004;20:2067–2074. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
