Erythropoietin receptor transcription is neither elevated nor predictive of surface expression in human tumour cells
- PMID: 18349818
- PMCID: PMC2275479
- DOI: 10.1038/sj.bjc.6604220
Erythropoietin receptor transcription is neither elevated nor predictive of surface expression in human tumour cells
Abstract
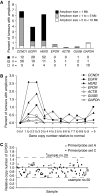
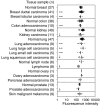
Erythropoietin receptor (EpoR) has been reported to be overexpressed in tumours and has raised safety concerns regarding the use of erythropoiesis-stimulating agents (ESAs) to treat anaemia in cancer patients. To investigate the potential for EpoR to be overexpressed in tumours, we have evaluated human tumours for amplification of the EPOR locus, levels of EPOR transcripts, and expression of surface EpoR protein. Gene amplification analysis of 1083 solid tumours found that amplification of the EPOR locus was rare with frequencies similar to other non-oncogenes. EPOR transcript levels in tumours and tumour cell lines were low in comparison with bone marrow and were equivalent to, or lower than, levels in normal tissues of tumour origin. Although EpoR mRNA was detected in some tumour lines, no EpoR could be detected on the cell surface using (125)I-Epo binding studies. This may be due to the lack of EpoR protein expression or lack of cell-surface-trafficking factors, such as Jak2. Taken together, we have found no evidence that EpoR is overexpressed in tumours or gets to the surface of tumour cells. This suggests that there is no selective advantage for tumours to overexpress EpoR and questions the functional relevance of EpoR gene transcription in tumours.
Figures



References
-
- Acs G, Acs P, Beckwith SM, Pitts RL, Clements E, Wong K, Verma A (2001) Erythropoietin and erythropoietin receptor expression in human cancer. Cancer Res 61: 3561–3565 - PubMed
-
- Acs G, Zhang PJ, McGrath CM, Acs P, McBroom J, Mohyeldin A, Liu S, Lu H, Verma A (2003) Hypoxia-inducible erythropoietin signaling in squamous dysplasia and squamous cell carcinoma of the uterine cervix and its potential role in cervical carcinogenesis and tumor progression. Am J Pathol 162: 1789–1806 - PMC - PubMed
-
- Berdel WE, Danhauser-Riedl S, Oberberg D, Zafferani M (1992) Effects of hematopoietic growth factors on malignant nonhematopoietic cells. Semin Oncol 19: 41–45 - PubMed
-
- Berdel WE, Oberberg D, Reufi B, Thiel E (1991) Studies on the role of recombinant human erythropoietin in the growth regulation of human nonhematopoietic tumor cells in vitro. Ann Hematol 63: 5–8 - PubMed
-
- Bohlius J, Wilson J, Seidenfeld J, Piper M, Schwarzer G, Sandercock J, Trelle S, Weingart O, Bayliss S, Djulbegovic B, Bennett CL, Langensiepen S, Hyde C, Engert A (2006) Recombinant human erythropoietins and cancer patients: updated meta-analysis of 57 studies including 9353 patients. J Natl Cancer Inst 98: 708–714 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

