Expression of the Drosophila secreted cuticle protein 73 (dsc73) requires Shavenbaby
- PMID: 18351665
- PMCID: PMC2826221
- DOI: 10.1002/dvdy.21512
Expression of the Drosophila secreted cuticle protein 73 (dsc73) requires Shavenbaby
Abstract
Low stringency genomic library screens with genomic fragments from the sex determination gene doublesex identified the Drosophila secreted cuticle protein 73 (dsc73) gene, which encodes an 852-residue protein with an N-terminal signal sequence. In embryos, dsc73 RNA and protein are expressed to high levels in the epidermal cells that secrete the larval cuticle as well as in other cuticle-secreting tissues such as the trachea and salivary duct. Embryonic expression of dsc73 requires Shavenbaby, a transcription factor regulating cuticle formation. Double-labeling experiments with alphaCrb and alphaSAS reveal that, as with chitin and other known cuticle proteins, Dsc73 is secreted apically. Zygotic loss of dsc73 results in larval lethality but loss does not result in overt patterning defects or overt morphological defects in the embryonic tissues in which it is expressed. Thus, dsc73 encodes a novel secreted protein, and it is conserved within the Drosophila group. dsc73 may serve as a useful embryonic marker for cuticular patterning.
(c) 2008 Wiley-Liss, Inc.
Figures


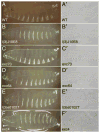
 , respectively, in a′ and a″). Although Dsc73 is expressed in the dorsal trunk cells of the trachea (DT), it is not detected in the dorsal branch (DB) or transverse connective (TC) (b′, b″, c′ and c″).
, respectively, in a′ and a″). Although Dsc73 is expressed in the dorsal trunk cells of the trachea (DT), it is not detected in the dorsal branch (DB) or transverse connective (TC) (b′, b″, c′ and c″).

References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Andrew DJ. PhD Thesis. UC; San Diego: 1987. A search for new genes regulating sex determination in Drosophila melanogaster.
-
- Baker BS, Wolfner MF. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes Dev. 1988;2:477–489. - PubMed
-
- Belote JM, Baker BS. The dual functions of a sex determination gene in Drosophila melanogaster. Dev Biol. 1983;95:512–517. - PubMed
-
- Britten RJ, Graham DE, Neufeld BR. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

