Generation of rho0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses
- PMID: 18353857
- PMCID: PMC2367725
- DOI: 10.1093/nar/gkn124
Generation of rho0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses
Abstract
Eukaryotic cells devoid of mitochondrial DNA (rho0 cells) were originally generated under artificial growth conditions utilizing ethidium bromide. The chemical is known to intercalate preferentially with the mitochondrial double-stranded DNA thereby interfering with enzymes of the replication machinery. Rho0 cell lines are highly valuable tools to study human mitochondrial disorders because they can be utilized in cytoplasmic transfer experiments. However, mutagenic effects of ethidium bromide onto the nuclear DNA cannot be excluded. To foreclose this mutagenic character during the development of rho0 cell lines, we developed an extremely mild, reliable and timesaving method to generate rho0 cell lines within 3-5 days based on an enzymatic approach. Utilizing the genes for the restriction endonuclease EcoRI and the fluorescent protein EGFP that were fused to a mitochondrial targeting sequence, we developed a CMV-driven expression vector that allowed the temporal expression of the resulting fusion enzyme in eukaryotic cells. Applied on the human cell line 143B.TK- the active protein localized to mitochondria and induced the complete destruction of endogenous mtDNA. Mouse and rat rho0 cell lines were also successfully created with this approach. Furthermore, the newly established 143B.TK- rho0 cell line was characterized in great detail thereby releasing interesting insights into the morphology and ultra structure of human rho0 mitochondria.
Figures

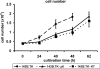
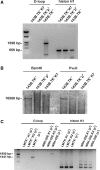
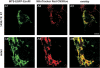



References
-
- Wallace DC. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found. Symp. 2001;235:247–263. - PubMed
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. - PubMed
-
- Borst P, Grivell LA. The mitochondrial genome of yeast. Cell. 1978;15:705–723. - PubMed
-
- Goldring ES, Grossman LI, Krupnick D, Cryer DR, Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J. Mol. Biol. 1970;52:323–335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

