Dissecting protein-RNA recognition sites
- PMID: 18353859
- PMCID: PMC2377425
- DOI: 10.1093/nar/gkn102
Dissecting protein-RNA recognition sites
Abstract
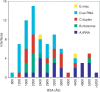
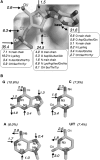
We analyze the protein-RNA interfaces in 81 transient binary complexes taken from the Protein Data Bank. Those with tRNA or duplex RNA are larger than with single-stranded RNA, and comparable in size to protein-DNA interfaces. The protein side bears a strong positive electrostatic potential and resembles protein-DNA interfaces in its amino acid composition. On the RNA side, the phosphate contributes less, and the sugar much more, to the interaction than in protein-DNA complexes. On average, protein-RNA interfaces contain 20 hydrogen bonds, 7 that involve the phosphates, 5 the sugar 2'OH, and 6 the bases, and 32 water molecules. The average H-bond density per unit buried surface area is less with tRNA or single-stranded RNA than with duplex RNA. The atomic packing is also less compact in interfaces with tRNA. On the protein side, the main chain NH and Arg/Lys side chains account for nearly half of all H-bonds to RNA; the main chain CO and side chain acceptor groups, for a quarter. The 2'OH is a major player in protein-RNA recognition, and shape complementarity an important determinant, whereas electrostatics and direct base-protein interactions play a lesser part than in protein-DNA recognition.
Figures



References
-
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, et al. The Protein Data Bank. Acta Crystallog. Sect. 2002;D58:899–907. - PubMed
-
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein–protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. - PubMed
-
- Nadassy K, Wodak S, Janin J. Structural features of protein–nucleic acid recognition sites. Biochemistry. 1999;38:1999–2017. - PubMed
-
- Jones S, van Heyningen P, Berman HM, Thornton JM. Protein-DNA interactions: a structural analysis. J. Mol. Biol. 1999;287:877–896. - PubMed

