Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS
- PMID: 18359832
- PMCID: PMC2394947
- DOI: 10.1128/AEM.00191-08
Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS
Abstract
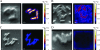
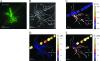
To examine phylogenetic identity and metabolic activity of individual cells in complex microbial communities, we developed a method which combines rRNA-based in situ hybridization with stable isotope imaging based on nanometer-scale secondary-ion mass spectrometry (NanoSIMS). Fluorine or bromine atoms were introduced into cells via 16S rRNA-targeted probes, which enabled phylogenetic identification of individual cells by NanoSIMS imaging. To overcome the natural fluorine and bromine backgrounds, we modified the current catalyzed reporter deposition fluorescence in situ hybridization (FISH) technique by using halogen-containing fluorescently labeled tyramides as substrates for the enzymatic tyramide deposition. Thereby, we obtained an enhanced element labeling of microbial cells by FISH (EL-FISH). The relative cellular abundance of fluorine or bromine after EL-FISH exceeded natural background concentrations by up to 180-fold and allowed us to distinguish target from non-target cells in NanoSIMS fluorine or bromine images. The method was optimized on single cells of axenic Escherichia coli and Vibrio cholerae cultures. EL-FISH/NanoSIMS was then applied to study interrelationships in a dual-species consortium consisting of a filamentous cyanobacterium and a heterotrophic alphaproteobacterium. We also evaluated the method on complex microbial aggregates obtained from human oral biofilms. In both samples, we found evidence for metabolic interactions by visualizing the fate of substrates labeled with (13)C-carbon and (15)N-nitrogen, while individual cells were identified simultaneously by halogen labeling via EL-FISH. Our novel approach will facilitate further studies of the ecophysiology of known and uncultured microorganisms in complex environments and communities.
Figures

References
-
- Adamczyk, J., M. Hesselsoe, N. Iversen, M. Horn, A. Lehner, P. H. Nielsen, M. Schloter, P. Roslev, and M. Wagner. 2003. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 69:6875-6887. - PMC - PubMed
-
- Blazejak, A., J. Kuever, C. Erseus, R. Amann, and N. Dubilier. 2006. Phylogeny of 16S rRNA, ribulose 1,5-bisphosphate carboxylase/oxygenase, and adenosine 5′-phosphosulfate reductase genes from gamma- and alphaproteobacterial symbionts in gutless marine worms (Oligochaeta) from Bermuda and the Bahamas. Appl. Environ. Microbiol. 72:5527-5536. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

