Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley
- PMID: 18359843
- PMCID: PMC2330320
- DOI: 10.1104/pp.108.116418
Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley
Abstract
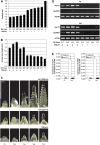
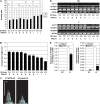
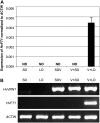
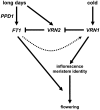
Interactions between flowering time genes were examined in a doubled haploid barley (Hordeum vulgare) population segregating for H. vulgare VERNALIZATION1 (HvVRN1), HvVRN2, and PHOTOPERIOD1 (PPD-H1). A deletion allele of HvVRN2 was associated with rapid inflorescence initiation and early flowering, but only in lines with an active allele of PPD-H1. In these lines, the floral promoter FLOWERING LOCUS T (HvFT1) was expressed at high levels without vernalization, and this preceded induction of HvVRN1. Lines with the deletion allele of HvVRN2 and the inactive ppd-H1 allele did not undergo rapid inflorescence initiation and were late flowering. These data suggest that HvVRN2 counteracts PPD-H1 to prevent flowering prior to vernalization. An allele of HvVRN1 that is expressed at high basal levels (HvVRN1-1) was associated with rapid inflorescence initiation regardless of HvVRN2 or PPD-H1 genotype. HvFT1 was expressed without vernalization in lines with the HvVRN1-1 allele and HvFT1 transcript levels were highest in lines with the active PPD-H1 allele; this correlated with rapid apex development postinflorescence initiation. Thus, expression of HvVRN1 promotes inflorescence initiation and up-regulates HvFT1. Analysis of HvVRN1 expression in different genetic backgrounds postvernalization showed that HvVRN2, HvFT1, and PPD-H1 are unlikely to play a role in low-temperature induction of HvVRN1. In a vernalization responsive barley, HvFT1 is not induced by low temperatures alone, but can be induced by long days following prolonged low-temperature treatment. We conclude that low-temperature and daylength flowering-response pathways are integrated to control expression of HvFT1 in barley, and that this might occur through regulation of HvVRN2 activity.
Figures




Similar articles
-
Major flowering time genes of barley: allelic diversity, effects, and comparison with wheat.Theor Appl Genet. 2021 Jul;134(7):1867-1897. doi: 10.1007/s00122-021-03824-z. Epub 2021 May 9. Theor Appl Genet. 2021. PMID: 33969431 Free PMC article. Review.
-
The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare).J Exp Bot. 2009;60(7):2169-78. doi: 10.1093/jxb/erp098. Epub 2009 Apr 8. J Exp Bot. 2009. PMID: 19357429 Free PMC article.
-
HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status.Plant Physiol. 2006 Apr;140(4):1397-405. doi: 10.1104/pp.105.073486. Epub 2006 Feb 24. Plant Physiol. 2006. PMID: 16500994 Free PMC article.
-
Fine-tuning of the flowering time control in winter barley: the importance of HvOS2 and HvVRN2 in non-inductive conditions.BMC Plant Biol. 2019 Mar 25;19(1):113. doi: 10.1186/s12870-019-1727-9. BMC Plant Biol. 2019. PMID: 30909882 Free PMC article.
-
The molecular basis of vernalization in different plant groups.Cold Spring Harb Symp Quant Biol. 2012;77:105-15. doi: 10.1101/sqb.2013.77.014449. Epub 2013 Apr 25. Cold Spring Harb Symp Quant Biol. 2012. PMID: 23619014 Review.
Cited by
-
Major flowering time genes of barley: allelic diversity, effects, and comparison with wheat.Theor Appl Genet. 2021 Jul;134(7):1867-1897. doi: 10.1007/s00122-021-03824-z. Epub 2021 May 9. Theor Appl Genet. 2021. PMID: 33969431 Free PMC article. Review.
-
A Vernalization Response in a Winter Safflower (Carthamus tinctorius) Involves the Upregulation of Homologs of FT, FUL, and MAF.Front Plant Sci. 2021 Mar 30;12:639014. doi: 10.3389/fpls.2021.639014. eCollection 2021. Front Plant Sci. 2021. PMID: 33859660 Free PMC article.
-
Advancing understanding of oat phenology for crop adaptation.Front Plant Sci. 2022 Oct 14;13:955623. doi: 10.3389/fpls.2022.955623. eCollection 2022. Front Plant Sci. 2022. PMID: 36311119 Free PMC article. Review.
-
Altered regulation of flowering expands growth ranges and maximizes yields in major crops.Front Plant Sci. 2023 Jan 19;14:1094411. doi: 10.3389/fpls.2023.1094411. eCollection 2023. Front Plant Sci. 2023. PMID: 36743503 Free PMC article. Review.
-
Flowering time runs hot and cold.Plant Physiol. 2022 Aug 29;190(1):5-18. doi: 10.1093/plphys/kiac111. Plant Physiol. 2022. PMID: 35274728 Free PMC article. Review.
References
-
- Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E (2006) The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 46 462–476 - PubMed
-
- Boyd WJR, Li CD, Grime CR, Cakir M, Potipibool S, Kaveeta L, Men S, Jamal-Kamali MR, Barr AR, Moody DB, et al (2003) Conventional and molecular genetic analysis of factors contributing to variation in the timing of heading among spring barley (Hordeum vulgare L.) genotypes grown over a mild winter growing season. Aust J Agric Res 54 1277–1301
-
- Chang S, Puryear J, Cairney K (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116
-
- Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18 675–689 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

