Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2
- PMID: 18359851
- PMCID: PMC2290789
- DOI: 10.1073/pnas.0712216105
Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2
Abstract
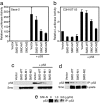
The hedgehog (Hh) signaling pathway regulates the development of many organs in mammals, and activation of this pathway is widely observed in human cancers. Although it is known that Hh signaling activates the expression of genes involved in cell growth, the precise role of the Hh pathway in cancer development is still unclear. Here, we show that constitutively activated mutants of Smoothened (Smo), a transducer of the Hh signaling pathway, inhibit the accumulation of the tumor suppressor protein p53. This inhibition was also observed in the presence of Hh ligand or with the overexpression of the transcription factors Gli1 and Gli2, downstream effectors of Smo, indicating that this inhibition is specific for the Hh pathway. We also report that Smo mutants augment p53 binding to the E3 ubiquitin-protein ligase Mdm2 and promote p53 ubiquitination. Furthermore, Hh signaling induced the phosphorylation of human Mdm2 protein on serines 166 and 186, which are activating phosphorylation sites of Mdm2. Smo mutants enhanced the proliferation of mouse embryonic fibroblasts (MEFs) while inducing a DNA-damage response. Moreover, Smo partially inhibited p53-dependent apoptosis and cell growth inhibition in oncogene-expressing MEFs. We also found that accumulation of p53 is inhibited by Hh signaling in several human cancer cell lines. Therefore, the Hh pathway may be a powerful accelerator of oncogenesis by activating cell proliferation and inhibiting the p53-mediated anti-cancer barrier induced by oncogenic stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


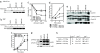

References
-
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. - PubMed
-
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15:3059–3087. - PubMed
-
- Ingham PW, Placzek M. Orchestrating ontogenesis: Variations on a theme by sonic hedgehog. Nat Rev Genet. 2006;7:841–850. - PubMed
-
- Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. - PubMed
-
- Berman DM, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

