The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions
- PMID: 18361515
- PMCID: PMC2839126
- DOI: 10.1021/pr700764j
The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions
Abstract
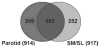
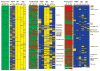
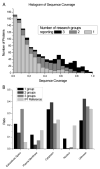
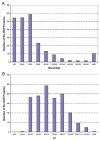
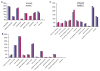
Saliva is a body fluid with important functions in oral and general health. A consortium of three research groups catalogued the proteins in human saliva collected as the ductal secretions: 1166 identifications--914 in parotid and 917 in submandibular/sublingual saliva--were made. The results showed that a high proportion of proteins that are found in plasma and/or tears are also present in saliva along with unique components. The proteins identified are involved in numerous molecular processes ranging from structural functions to enzymatic/catalytic activities. As expected, the majority mapped to the extracellular and secretory compartments. An immunoblot approach was used to validate the presence in saliva of a subset of the proteins identified by mass spectrometric approaches. These experiments focused on novel constituents and proteins for which the peptide evidence was relatively weak. Ultimately, information derived from the work reported here and related published studies can be used to translate blood-based clinical laboratory tests into a format that utilizes saliva. Additionally, a catalogue of the salivary proteome of healthy individuals allows future analyses of salivary samples from individuals with oral and systemic diseases, with the goal of identifying biomarkers with diagnostic and/or prognostic value for these conditions; another possibility is the discovery of therapeutic targets.
Figures
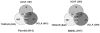


References
-
- Levine MJ. Ann NY Acad Sci. 1993;694:11–16. - PubMed
-
- Lamkin MS, Oppenheim FG. Crit Rev Oral Biol Med. 1993;4:251–259. - PubMed
-
- Mandel ID. J Dent Res. 1987;66:623–627. - PubMed
-
- Humphrey SP, Williamson RT. J Prosthet Dent. 2001;85:162–169. - PubMed
-
- Dodds MW, Johnson DA, Yeh CK. J Dent. 2005;33:223–233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

