Moraxella catarrhalis expresses an unusual Hfq protein
- PMID: 18362134
- PMCID: PMC2423088
- DOI: 10.1128/IAI.01652-07
Moraxella catarrhalis expresses an unusual Hfq protein
Abstract
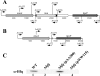
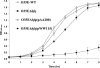
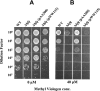
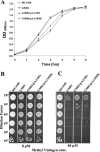
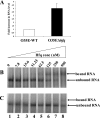
The Hfq protein is recognized as a global regulatory molecule that facilitates certain RNA-RNA interactions in bacteria. BLAST analysis identified a 630-nucleotide open reading frame in the genome of Moraxella catarrhalis ATCC 43617 that was highly conserved among M. catarrhalis strains and which encoded a predicted protein with significant homology to the Hfq protein of Escherichia coli. This protein, containing 210 amino acids, was more than twice as large as the Hfq proteins previously described for other bacteria. The C-terminal half of the M. catarrhalis Hfq protein was very hydrophilic and contained two different types of amino acid repeats. A mutation in the M. catarrhalis hfq gene affected both the growth rate of this organism and its sensitivity to at least two different types of stress in vitro. Provision of the wild-type M. catarrhalis hfq gene in trans eliminated these phenotypic differences in the hfq mutant. This M. catarrhalis hfq mutant exhibited altered expression of some cell envelope proteins relative to the wild-type parent strain and also had a growth advantage in a continuous flow biofilm system. The presence of the wild-type M. catarrhalis hfq gene in trans in an E. coli hfq mutant fully reversed the modest growth deficiency of this E. coli mutant and partially reversed the stress sensitivity of this E. coli mutant to methyl viologen. The use of an electrophoretic mobility shift assay showed that this M. catarrhalis Hfq protein could bind RNA derived from a gene whose expression was altered in the M. catarrhalis hfq mutant.
Figures



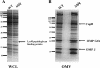

Similar articles
-
Moraxella catarrhalis synthesizes an autotransporter that is an acid phosphatase.J Bacteriol. 2008 Feb;190(4):1459-72. doi: 10.1128/JB.01688-07. Epub 2007 Dec 7. J Bacteriol. 2008. PMID: 18065547 Free PMC article.
-
Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites.Biochemistry. 2006 Apr 18;45(15):4875-87. doi: 10.1021/bi0523613. Biochemistry. 2006. PMID: 16605255
-
Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi.Mol Microbiol. 2010 Nov;78(3):622-35. doi: 10.1111/j.1365-2958.2010.07374.x. Epub 2010 Sep 27. Mol Microbiol. 2010. PMID: 20815822 Free PMC article.
-
A Moraxella catarrhalis two-component signal transduction system necessary for growth in liquid media affects production of two lysozyme inhibitors.Infect Immun. 2015 Jan;83(1):146-60. doi: 10.1128/IAI.02486-14. Epub 2014 Oct 13. Infect Immun. 2015. PMID: 25312959 Free PMC article.
-
The RNA chaperone Hfq is important for growth and stress tolerance in Francisella novicida.PLoS One. 2011 May 5;6(5):e19797. doi: 10.1371/journal.pone.0019797. PLoS One. 2011. PMID: 21573133 Free PMC article.
Cited by
-
The RNA chaperone Hfq promotes fitness of Actinobacillus pleuropneumoniae during porcine pleuropneumonia.Infect Immun. 2013 Aug;81(8):2952-61. doi: 10.1128/IAI.00392-13. Epub 2013 Jun 3. Infect Immun. 2013. PMID: 23732171 Free PMC article.
-
Insights into the complementation potential of the extreme acidophile's orthologue in replacing Escherichia coli hfq gene-particularly in bacterial resistance to environmental stress.World J Microbiol Biotechnol. 2024 Feb 22;40(4):105. doi: 10.1007/s11274-024-03924-0. World J Microbiol Biotechnol. 2024. PMID: 38386219
-
Clostridium difficile Hfq can replace Escherichia coli Hfq for most of its function.RNA. 2014 Oct;20(10):1567-78. doi: 10.1261/rna.043372.113. Epub 2014 Aug 21. RNA. 2014. PMID: 25147238 Free PMC article.
-
Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora.J Bacteriol. 2013 Apr;195(8):1706-17. doi: 10.1128/JB.02056-12. Epub 2013 Feb 1. J Bacteriol. 2013. PMID: 23378513 Free PMC article.
-
Acidic C-terminal domains autoregulate the RNA chaperone Hfq.Elife. 2017 Aug 9;6:e27049. doi: 10.7554/eLife.27049. Elife. 2017. PMID: 28826489 Free PMC article.
References
-
- Adlowitz, D. G., T. Hiltke, A. J. Lesse, and T. F. Murphy. 2004. Identification and characterization of outer membrane proteins G1a and G1b of Moraxella catarrhalis. Vaccine 222533-2540. - PubMed
-
- Adlowitz, D. G., C. Kirkham, S. Sethi, and T. F. Murphy. 2006. Human serum and mucosal antibody responses to outer membrane protein G1b of Moraxella catarrhalis in chronic obstructive pulmonary disease. FEMS Immunol. Med. Microbiol. 46139-146. - PubMed
-
- Aebi, C., B. Stone, M. Beucher, L. D. Cope, I. Maciver, S. E. Thomas, G. H. McCracken, Jr., P. F. Sparling, and E. J. Hansen. 1996. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect. Immun. 642024-2030. - PMC - PubMed
-
- Arluison, V., P. Derreumaux, F. Allemand, M. Folichon, E. Hajnsdorf, and P. Regnier. 2002. Structural modelling of the Sm-like protein Hfq from Escherichia coli. J. Mol. Biol. 320705-712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

