Mapping of POP1-binding site on pyrin domain of ASC
- PMID: 18362139
- PMCID: PMC3258898
- DOI: 10.1074/jbc.M801589200
Mapping of POP1-binding site on pyrin domain of ASC
Abstract
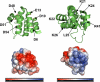
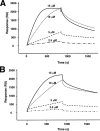
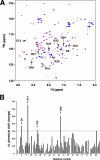
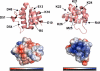
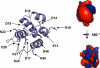
Apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) is an essential adaptor protein in the formation of a multiprotein complex that activates procaspase-1. ASC is also known as a modulator of NF-kappaB activation pathways. ASC has a bipartite domain structure, consisting of an N-terminal pyrin domain (PYD) and a C-terminal caspase-recruitment domain. The PYD of ASC (ASC_PYD) is known to interact with various PYD-containing intracellular danger signal sensors and PYD-only proteins. Using purified proteins, we characterized the in vitro interaction of ASC_PYD with PYD-only protein 1 (POP1). POP1 specifically interacts with ASC_PYD with a dissociation constant of 4.08 +/- 0.52 microm but does not interact with Cryopyrin. NMR and mutagenesis experiments show that a negative electrostatic potential surface patch (EPSP) on ASC_PYD, consisting of the first (H1) and fourth (H4) helices, is essential in the interaction with POP1. A positive EPSP on POP1, consisting of the second (H2) and third (H3) helices, is a counterpart of this interaction. The interaction between ASC_PYD and POP1 is similar to the interaction between caspase recruitment domains of Apaf-1 and procaspase-9. In addition, we present evidence that conformational changes at the long loop of ASC_PYD between the H2 and H3 helices can affect its interaction with POP1. Based on our observations, we propose that the positive EPSP of ASC_PYD, including the H2 and H3 helices, may be the binding site for Cryopyrin, and the interaction with Cryopyrin may induce the dissociation of POP1 from ASC_PYD.
Figures



References
-
- Masumoto, J., Taniguchi, S., Ayukawa, K., Sarvotham, H., Kishino, T., Niikawa, N., Hidaka, E., Katsuyama, T., Higuchi, T., and Sagara, J. (1999) J. Biol. Chem. 274 33835-33838 - PubMed
-
- McConnell, B. B., and Vertino, P. M. (2004) Apoptosis 9 5-18 - PubMed
-
- Srinivasula, S. M., Poyet, J. L., Razmara, M., Datta, P., Zhang, Z., and Alnemri, E. S. (2002) J. Biol. Chem. 277 21119-21122 - PubMed
-
- Martinon, F., and Tschopp, J. (2007) Cell Death Differ. 14 10-22 - PubMed
-
- Agostini, L., Martinon, F., Burns, K., McDermott, M. F., Hawkins, P. N., and Tschopp, J. (2004) Immunity 20 319-325 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

