Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response
- PMID: 18362164
- PMCID: PMC2423288
- DOI: 10.1128/MCB.02284-07
Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response
Abstract
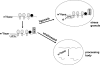
Nonsense-mediated RNA decay (NMD) rapidly degrades both mutated mRNAs and nonmutated cellular mRNAs in what is thought to be a constitutive fashion. Here we demonstrate that NMD is inhibited in hypoxic cells and that this inhibition is dependent on phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (eIF2alpha). eIF2alpha phosphorylation is known to promote translational and transcriptional up-regulation of genes important for the cellular response to stress. We show that the mRNAs of several of these stress-induced genes are NMD targets and that the repression of NMD stabilizes these mRNAs, thus demonstrating that the inhibition of NMD augments the cellular stress response. Furthermore, hypoxia-induced formation of cytoplasmic stress granules is also dependent on eIF2alpha phosphorylation, and components of the NMD pathway are relocalized to these granules in hypoxic cells, providing a potential mechanism for the hypoxic inhibition of NMD. Our demonstration that NMD is inhibited in hypoxic cells reveals that the regulation of NMD can dynamically alter gene expression and also establishes a novel mechanism for hypoxic gene regulation.
Figures
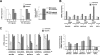
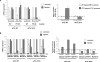
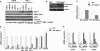


References
-
- Bi, M., C. Naczki, M. Koritzinsky, D. Fels, J. Blais, N. Hu, H. Harding, I. Novoa, M. Varia, J. Raleigh, D. Scheuner, R. J. Kaufman, J. Bell, D. Ron, B. G. Wouters, and C. Koumenis. 2005. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 243470-3481. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
