Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger
- PMID: 18364712
- PMCID: PMC2290933
- DOI: 10.1038/msb.2008.12
Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger
Abstract
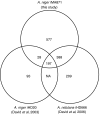
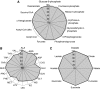
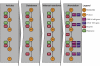
The release of the genome sequences of two strains of Aspergillus niger has allowed systems-level investigations of this important microbial cell factory. To this end, tools for doing data integration of multi-ome data are necessary, and especially interesting in the context of metabolism. On the basis of an A. niger bibliome survey, we present the largest model reconstruction of a metabolic network reported for a fungal species. The reconstructed gapless metabolic network is based on the reportings of 371 articles and comprises 1190 biochemically unique reactions and 871 ORFs. Inclusion of isoenzymes increases the total number of reactions to 2240. A graphical map of the metabolic network is presented. All levels of the reconstruction process were based on manual curation. From the reconstructed metabolic network, a mathematical model was constructed and validated with data on yields, fluxes and transcription. The presented metabolic network and map are useful tools for examining systemwide data in a metabolic context. Results from the validated model show a great potential for expanding the use of A. niger as a high-yield production platform.
Figures





References
-
- Baker SE (2006) Aspergillus niger genomics: past, present and into the future. Med Mycol 44: S17–S21 - PubMed
-
- Brennan PJ, Griffin PFS, Lösel DM, Tyrrell D (1974) The lipids of fungi. Prog Chem Fats Other Lipids 14: 49–89 - PubMed
-
- Byrne PF, Brennan PJ (1976) Isolation and characterization of inositol-containing glycosphingolipids from Aspergillus niger. Biochem Soc Trans 4: 893–895 - PubMed
-
- Chattopadhyay P, Banerjee SK, Sen K, Chakrabarti P (1985) Lipid profiles of Aspergillus niger and its unsaturated fatty acid auxotroph, UFA2. Can J Microbiol 31: 352–355 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

