Genome-wide detection of serpentine receptor-like proteins in malaria parasites
- PMID: 18365025
- PMCID: PMC2268965
- DOI: 10.1371/journal.pone.0001889
Genome-wide detection of serpentine receptor-like proteins in malaria parasites
Abstract
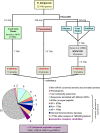
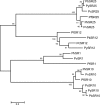
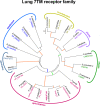
Serpentine receptors comprise a large family of membrane receptors distributed over diverse organisms, such as bacteria, fungi, plants and all metazoans. However, the presence of serpentine receptors in protozoan parasites is largely unknown so far. In the present study we performed a genome-wide search for proteins containing seven transmembrane domains (7-TM) in the human malaria parasite Plasmodium falciparum and identified four serpentine receptor-like proteins. These proteins, denoted PfSR1, PfSR10, PfSR12 and PfSR25, show membrane topologies that resemble those exhibited by members belonging to different families of serpentine receptors. Expression of the pfsrs genes was detected by Real Time PCR in P. falciparum intraerythrocytic stages, indicating that they potentially code for functional proteins. We also found corresponding homologues for the PfSRs in five other Plasmodium species, two primate and three rodent parasites. PfSR10 and 25 are the most conserved receptors among the different species, while PfSR1 and 12 are more divergent. Interestingly, we found that PfSR10 and PfSR12 possess similarity to orphan serpentine receptors of other organisms. The identification of potential parasite membrane receptors raises a new perspective for essential aspects of malaria parasite host cell infection.
Conflict of interest statement
Figures

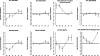
References
-
- Bannister L, Mitchell G. The ins, outs and roundabouts of malaria. Trends Parasitol. 2003;19:209–13. - PubMed
-
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–8. - PubMed
-
- Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

