A multi-template combination algorithm for protein comparative modeling
- PMID: 18366648
- PMCID: PMC2311309
- DOI: 10.1186/1472-6807-8-18
A multi-template combination algorithm for protein comparative modeling
Abstract
Background: Multiple protein templates are commonly used in manual protein structure prediction. However, few automated algorithms of selecting and combining multiple templates are available.
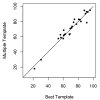
Results: Here we develop an effective multi-template combination algorithm for protein comparative modeling. The algorithm selects templates according to the similarity significance of the alignments between template and target proteins. It combines the whole template-target alignments whose similarity significance score is close to that of the top template-target alignment within a threshold, whereas it only takes alignment fragments from a less similar template-target alignment that align with a sizable uncovered region of the target. We compare the algorithm with the traditional method of using a single top template on the 45 comparative modeling targets (i.e. easy template-based modeling targets) used in the seventh edition of Critical Assessment of Techniques for Protein Structure Prediction (CASP7). The multi-template combination algorithm improves the GDT-TS scores of predicted models by 6.8% on average. The statistical analysis shows that the improvement is significant (p-value < 10-4). Compared with the ideal approach that always uses the best template, the multi-template approach yields only slightly better performance. During the CASP7 experiment, the preliminary implementation of the multi-template combination algorithm (FOLDpro) was ranked second among 67 servers in the category of high-accuracy structure prediction in terms of GDT-TS measure.
Conclusion: We have developed a novel multi-template algorithm to improve protein comparative modeling.
Figures




References
-
- Vitkup D, Melamud E, Moult J, Sander C. Completeness in structural genomics. Nature Struct Biol. 2001;8:559–566. - PubMed
-
- Brenner S. A tour of structural genomics. Nature Rev Genet. 2001;2:801–809. - PubMed
-
- Browne W, North A, Philips D, Brew K, Vanaman T, Hill R. A possible three-dimensional structure of bovine alpha-lactalbumin based on that of hen.s egg-white lysozyme. J Mol Biol. 1969;42:65–86. - PubMed
-
- Blundell T, Sibanda B, Sternberg M, Thornton J. Knowledge-based prediction of protein structures and the design of novel molecules. Nature. 1987;326:347–352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

