SNP selection for genes of iron metabolism in a study of genetic modifiers of hemochromatosis
- PMID: 18366708
- PMCID: PMC2289803
- DOI: 10.1186/1471-2350-9-18
SNP selection for genes of iron metabolism in a study of genetic modifiers of hemochromatosis
Abstract
Background: We report our experience of selecting tag SNPs in 35 genes involved in iron metabolism in a cohort study seeking to discover genetic modifiers of hereditary hemochromatosis.
Methods: We combined our own and publicly available resequencing data with HapMap to maximise our coverage to select 384 SNPs in candidate genes suitable for typing on the Illumina platform.
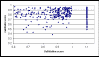
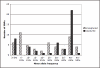
Results: Validation/design scores above 0.6 were not strongly correlated with SNP performance as estimated by Gentrain score. We contrasted results from two tag SNP selection algorithms, LDselect and Tagger. Varying r2 from 0.5 to 1.0 produced a near linear correlation with the number of tag SNPs required. We examined the pattern of linkage disequilibrium of three levels of resequencing coverage for the transferrin gene and found HapMap phase 1 tag SNPs capture 45% of the > or = 3% MAF SNPs found in SeattleSNPs where there is nearly complete resequencing. Resequencing can reveal adjacent SNPs (within 60 bp) which may affect assay performance. We report the number of SNPs present within the region of six of our larger candidate genes, for different versions of stock genotyping assays.
Conclusion: A candidate gene approach should seek to maximise coverage, and this can be improved by adding to HapMap data any available sequencing data. Tag SNP software must be fast and flexible to data changes, since tag SNP selection involves iteration as investigators seek to satisfy the competing demands of coverage within and between populations, and typability on the technology platform chosen.
Figures



References
-
- Consortium TIHM. The International HapMap Project. Nature. 2003;426:789–796. - PubMed
-
- Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Jr., Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. - PubMed
-
- Giles GG, English DR. The Melbourne Collaborative Cohort Study. IARC Sci Publ. 2002;156:69–70. - PubMed
-
- McLaren CE, Barton JC, Adams PC, Harris EL, Acton RT, Press N, Reboussin DM, McLaren GD, Sholinsky P, Walker AP, Gordeuk VR, Leiendecker-Foster C, Dawkins FW, Eckfeldt JH, Mellen BG, Speechley M, Thomson E. Hemochromatosis and Iron Overload Screening (HEIRS) study design for an evaluation of 100,000 primary care-based adults. Am J Med Sci. 2003;325:53–62. - PubMed

