Comparative genomic analysis of Mycobacterium avium subspecies obtained from multiple host species
- PMID: 18366709
- PMCID: PMC2323391
- DOI: 10.1186/1471-2164-9-135
Comparative genomic analysis of Mycobacterium avium subspecies obtained from multiple host species
Abstract
Background: Mycobacterium avium (M. avium) subspecies vary widely in both pathogenicity and host specificity, but the genetic features contributing to this diversity remain unclear.
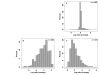
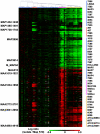
Results: A comparative genomic approach was used to identify large sequence polymorphisms among M. avium subspecies obtained from a variety of host animals. DNA microarrays were used as a platform for comparing mycobacterial isolates with the sequenced bovine isolate M. avium subsp. paratuberculosis (MAP) K-10. Open reading frames (ORFs) were classified as present or divergent based on the relative fluorescent intensities of the experimental samples compared to MAP K-10 DNA. Multiple large polymorphic regions were found in the genomes of MAP isolates obtained from sheep. One of these clusters encodes glycopeptidolipid biosynthesis enzymes which have not previously been identified in MAP. M. avium subsp. silvaticum isolates were observed to have a hybridization profile very similar to yet distinguishable from M. avium subsp. avium. Isolates obtained from cattle (n = 5), birds (n = 4), goats (n = 3), bison (n = 3), and humans (n = 9) were indistinguishable from cattle isolate MAP K-10.
Conclusion: Genome diversity in M. avium subspecies appears to be mediated by large sequence polymorphisms that are commonly associated with mobile genetic elements. Subspecies and host adapted isolates of M. avium were distinguishable by the presence or absence of specific polymorphisms.
Figures


References
-
- Saxegaard F, Baess I. Relationship between Mycobacterium avium, Mycobacterium paratuberculosis and "wood pigeon mycobacteria". Determinations by DNA-DNA hybridization. Apmis. 1988;96:37–42. - PubMed
-
- Thorel MF, Krichevsky M, Levy-Frebault VV. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int J Syst Bacteriol. 1990;40:254–260. - PubMed
-
- Paustian ML, Kapur V, Bannantine JP. Comparative genomic hybridizations reveal genetic regions within the Mycobacterium avium complex that are divergent from Mycobacterium avium subsp. paratuberculosis isolates. J Bacteriol. 2005;187:2406–2415. doi: 10.1128/JB.187.7.2406-2415.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

