OpenMS - an open-source software framework for mass spectrometry
- PMID: 18366760
- PMCID: PMC2311306
- DOI: 10.1186/1471-2105-9-163
OpenMS - an open-source software framework for mass spectrometry
Abstract
Background: Mass spectrometry is an essential analytical technique for high-throughput analysis in proteomics and metabolomics. The development of new separation techniques, precise mass analyzers and experimental protocols is a very active field of research. This leads to more complex experimental setups yielding ever increasing amounts of data. Consequently, analysis of the data is currently often the bottleneck for experimental studies. Although software tools for many data analysis tasks are available today, they are often hard to combine with each other or not flexible enough to allow for rapid prototyping of a new analysis workflow.
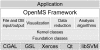
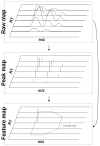
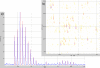
Results: We present OpenMS, a software framework for rapid application development in mass spectrometry. OpenMS has been designed to be portable, easy-to-use and robust while offering a rich functionality ranging from basic data structures to sophisticated algorithms for data analysis. This has already been demonstrated in several studies.
Conclusion: OpenMS is available under the Lesser GNU Public License (LGPL) from the project website at http://www.openms.de.
Figures

References
-
- Waters Corporation – MassLynx http://www.waters.com
-
- GE Healthcare – DeCyder http://www.gelifesciences.com
-
- Bellew M, Coram M, Fitzgibbon M, Igra M, Randolph T, Wang P, May D, Eng J, Fang R, Lin CW, Chen J, Goodlett D, Whiteaker J, Paulovich A, McIntosh M. A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics. 2006;22:1902–1909. doi: 10.1093/bioinformatics/btl276. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

